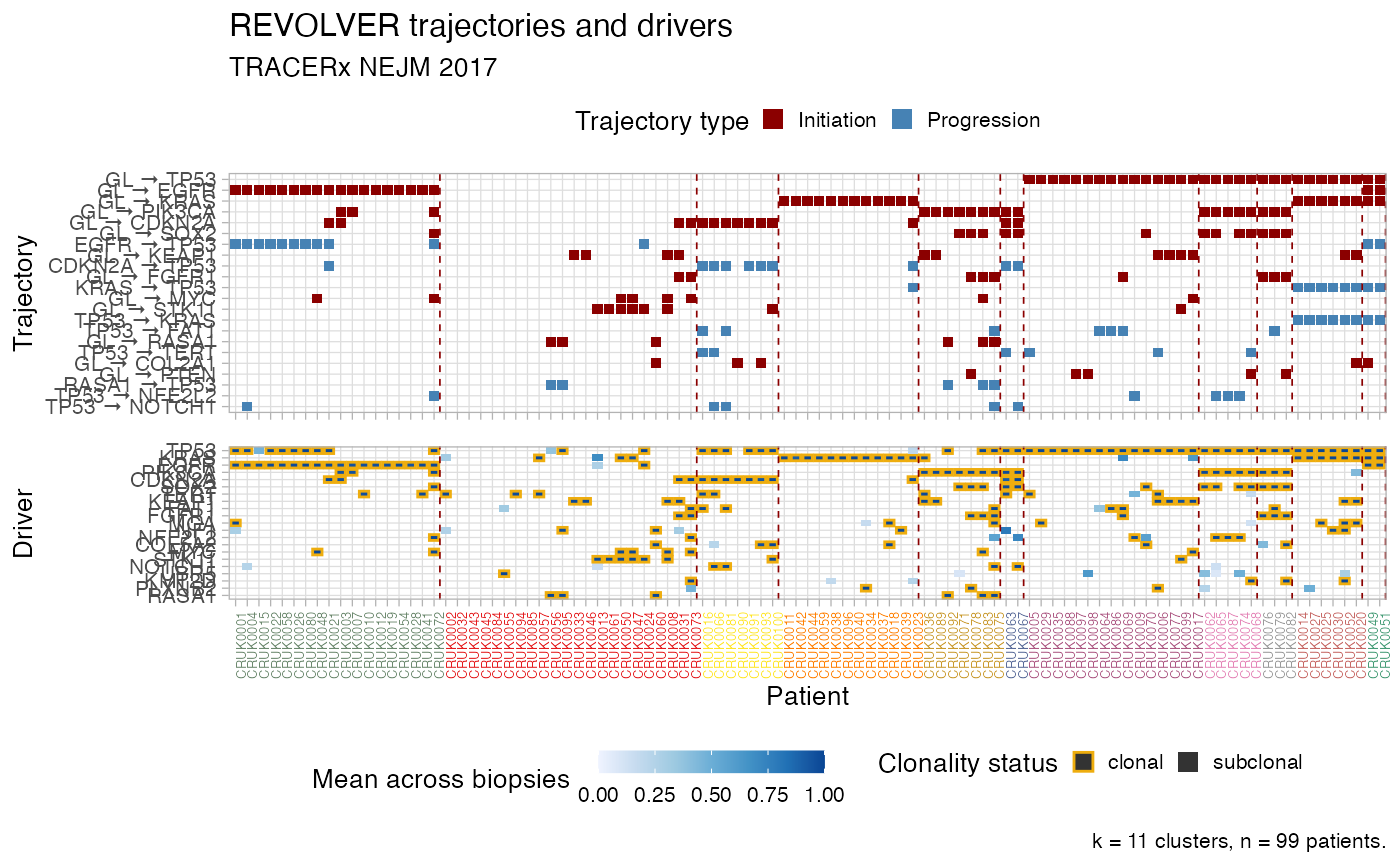
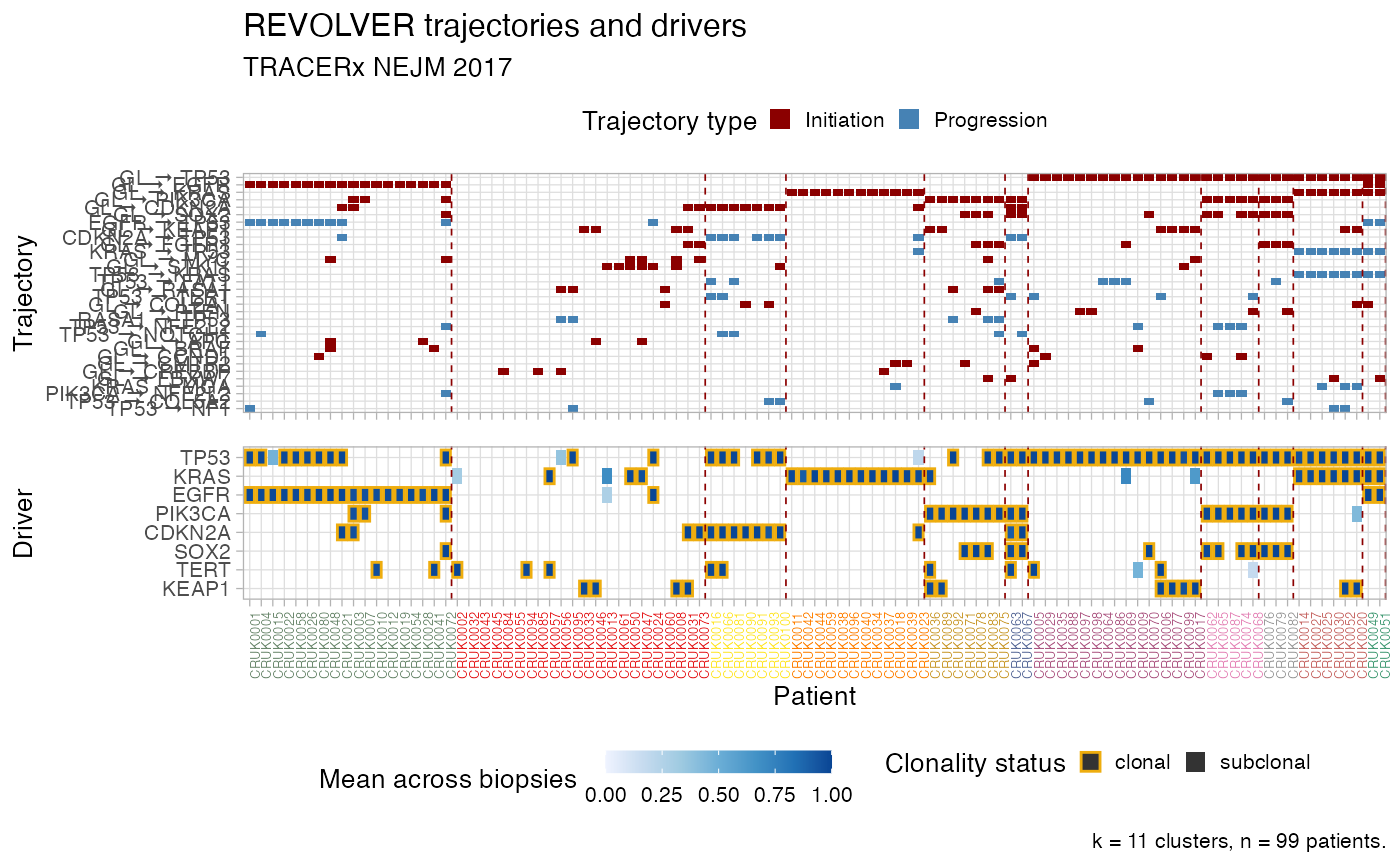
Plot the heatmaps of REVOLVER"s clusters, as tiles.
The top tile is patients vs trajectories, and bottom is patients vs drivers. For drivers colours reflect mean CCF/ binary values of a driver in every patient, and clonality status. For trajectories colours reflect if they are initiating or progressing, depending on the present of GL in the trajectory.
Patients are sorted by cluster to match the dendrogram that one can obtain with plot_dendrogram.
plot_clusters( x, cluster_palette = distinct_palette_few, cutoff_drivers = 5, cutoff_trajectories = 4, arrow.symbol = " → " )
Arguments
| x | A |
|---|---|
| cutoff_drivers | Plot only drivers that occur in at least |
| cutoff_trajectories | Plot only trajectories that occur in at least |
| arrow.symbol | UNICODE code to display arrows. Saving to PDF outputs with standard
methods (ggsave, cairo, etc), often can lead to errors with UNICODE chars; therefore either print to PNG or change
this variable to, e.g., |
| clusters_palette | A palette function that should return the colour of an arbitrary number of clusters. |
Value
A ggplot plot.
See also
Other Plotting functions:
distinct_palette_few(),
distinct_palette_many(),
gradient_palette(),
plot_DET_index(),
plot_dendrogram(),
plot_drivers_clonality(),
plot_drivers_graph(),
plot_drivers_occurrence(),
plot_jackknife_cluster_stability(),
plot_jackknife_coclustering(),
plot_jackknife_trajectories_stability(),
plot_patient_CCF_histogram(),
plot_patient_data(),
plot_patient_mutation_burden(),
plot_patient_oncoprint(),
plot_patient_trees_scores()
Examples
# Data released in the 'evoverse.datasets' data('TRACERx_NEJM_2017_REVOLVER', package = 'evoverse.datasets') plot_clusters(TRACERx_NEJM_2017_REVOLVER)#> Warning: Vectorized input to `element_text()` is not officially supported. #> Results may be unexpected or may change in future versions of ggplot2.plot_clusters(TRACERx_NEJM_2017_REVOLVER, cutoff_drivers = 10, cutoff_trajectories = 3)#> Warning: Vectorized input to `element_text()` is not officially supported. #> Results may be unexpected or may change in future versions of ggplot2.