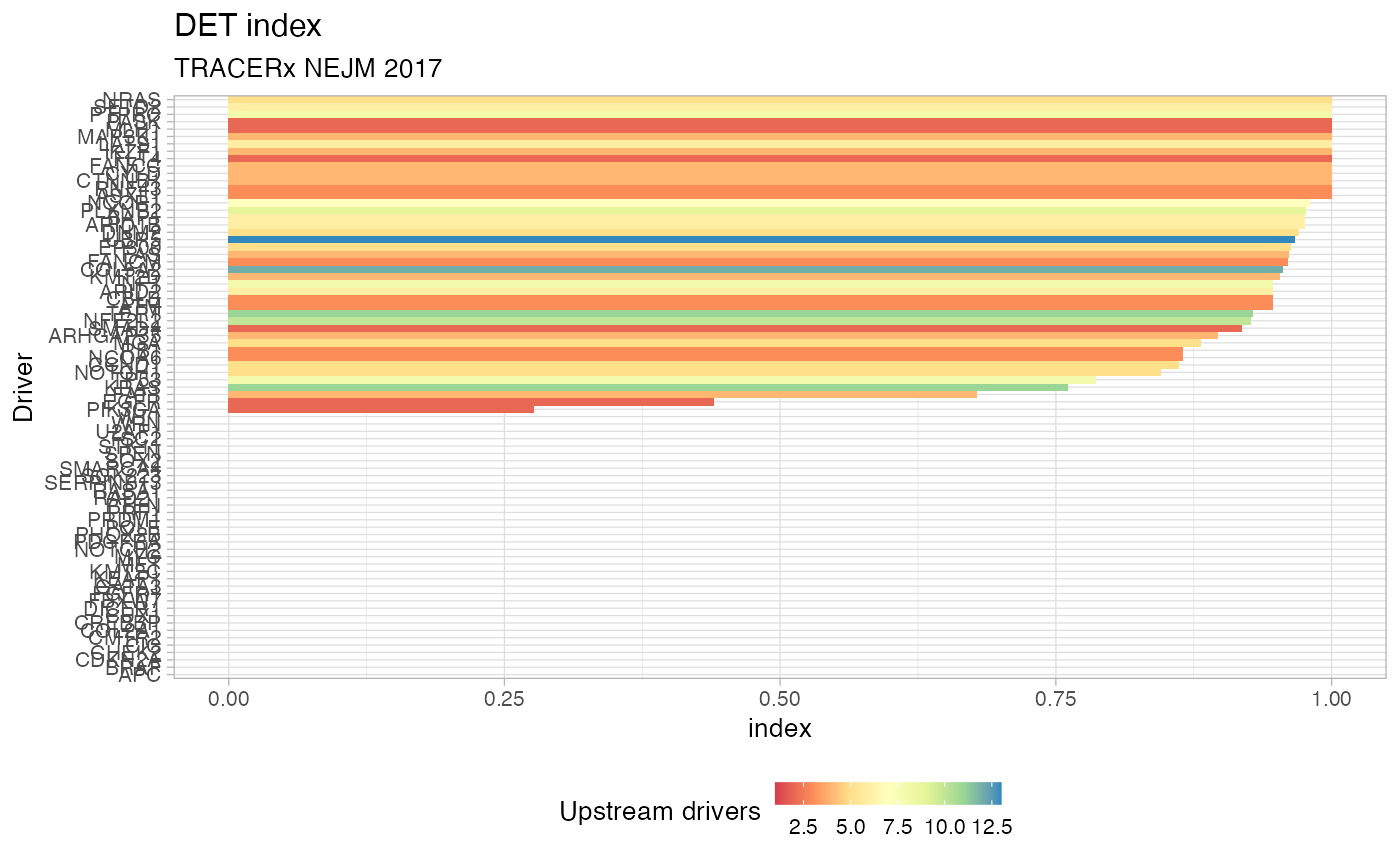
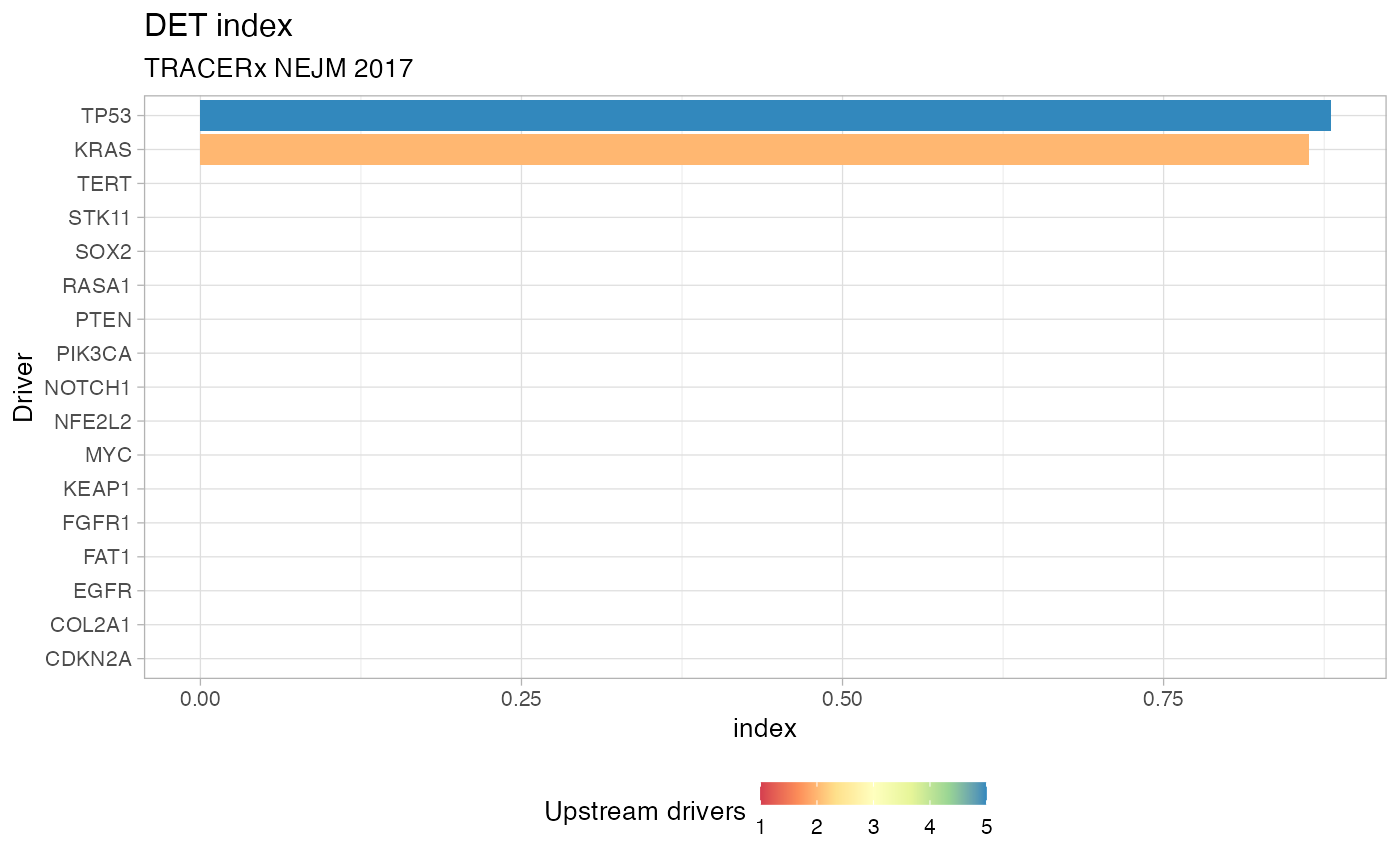
Plot the index of Divergent Evolutionary Trajectories, for a set of drivers
using function DET_index. The plot is a barplot with colours reflecting
the number of distinct incoming edges in each driver (species), and the height
reflecting the actual DET index value.
plot_DET_index(x, ...)
Arguments
| x | A |
|---|---|
| ... | Parmeters passed to function |
Value
A `ggplot` object of the plot.
See also
Other Plotting functions:
distinct_palette_few(),
distinct_palette_many(),
gradient_palette(),
plot_clusters(),
plot_dendrogram(),
plot_drivers_clonality(),
plot_drivers_graph(),
plot_drivers_occurrence(),
plot_jackknife_cluster_stability(),
plot_jackknife_coclustering(),
plot_jackknife_trajectories_stability(),
plot_patient_CCF_histogram(),
plot_patient_data(),
plot_patient_mutation_burden(),
plot_patient_oncoprint(),
plot_patient_trees_scores()
Examples
# Data released in the 'evoverse.datasets' data('TRACERx_NEJM_2017_REVOLVER', package = 'evoverse.datasets') plot_DET_index(TRACERx_NEJM_2017_REVOLVER)#> # A tibble: 79 x 4 #> driver diversity N DET_index #> <chr> <dbl> <int> <dbl> #> 1 APC 0 1 0 #> 2 BRAF 0 1 0 #> 3 CDKN2A 0 1 0 #> 4 CHEK2 0 1 0 #> 5 CIC 0 1 0 #> 6 CMTR2 0 1 0 #> 7 COL2A1 0 1 0 #> 8 CREBBP 0 1 0 #> 9 CUX1 0 1 0 #> 10 DICER1 0 1 0 #> # … with 69 more rows# Passing parameters to DET_index plot_DET_index(TRACERx_NEJM_2017_REVOLVER, min.occurrences = 5)#> # A tibble: 17 x 4 #> driver diversity N DET_index #> <chr> <dbl> <int> <dbl> #> 1 CDKN2A 0 1 0 #> 2 COL2A1 0 1 0 #> 3 EGFR 0 1 0 #> 4 FAT1 0 1 0 #> 5 FGFR1 0 1 0 #> 6 KEAP1 0 1 0 #> 7 MYC 0 1 0 #> 8 NFE2L2 0 1 0 #> 9 NOTCH1 0 1 0 #> 10 PIK3CA 0 1 0 #> 11 PTEN 0 1 0 #> 12 RASA1 0 1 0 #> 13 SOX2 0 1 0 #> 14 STK11 0 1 0 #> 15 TERT 0 1 0 #> 16 KRAS 0.598 2 0.863 #> 17 TP53 1.42 5 0.880