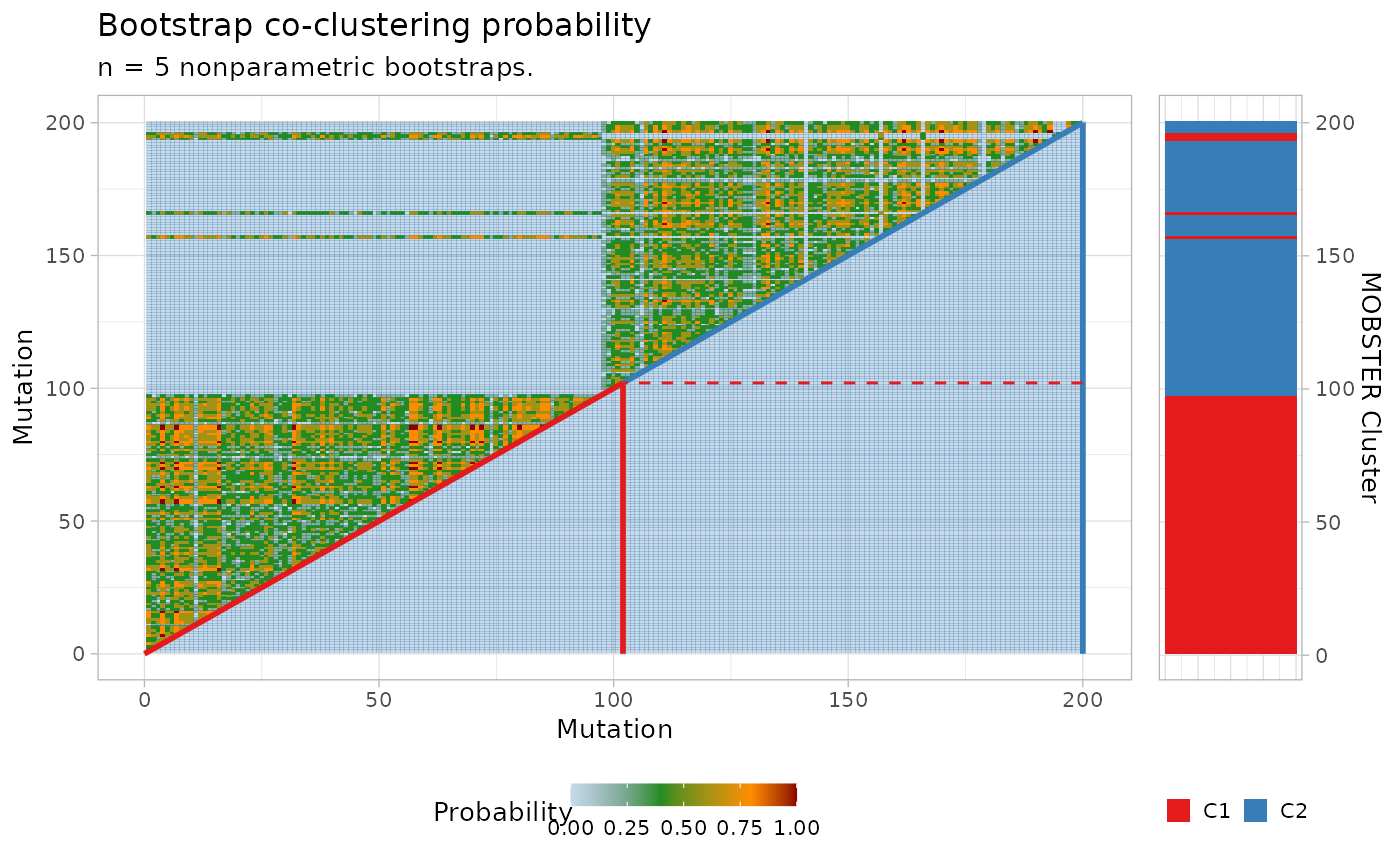
Plot the boostrapped co-clustering probability.
Source:R/plot_boostrap_coclustering.R
plot_bootstrap_coclustering.RdFrom the results of a call to mobster_boostrap, and
the results of a call to bootstrapped_statistics, a figure (heatmap-style)
for the boostrapped co-clustering probability is produced assembling plots
with cowplot::plot_grid.
Usage
plot_bootstrap_coclustering(
x,
bootstrap_results,
bootstrap_statistics,
colors = c(Tail = "gainsboro")
)Arguments
- x
A MOBSTER fit.
- bootstrap_results
Results of a call to
mobster_boostrap.- colors
If provided, these colours will be used for each cluster. If a subset of colours is provided, palette Set1 from
RColorBreweris used. By default the tail colour is provided as 'gainsboro'.- bootstrap_statisticsResults
of a call to
bootstrapped_statistics.
Examples
# Random small dataset
dataset = random_dataset(N = 200, seed = 123, Beta_variance_scaling = 100)
x = mobster_fit(dataset$data, auto_setup = 'FAST')
#> [ MOBSTER fit ]
#>
#> ✔ Loaded input data, n = 200.
#> ❯ n = 200. Mixture with k = 1,2 Beta(s). Pareto tail: TRUE and FALSE. Output
#> clusters with π > 0.02 and n > 10.
#> ! mobster automatic setup FAST for the analysis.
#> ❯ Scoring (without parallel) 2 x 2 x 2 = 8 models by reICL.
#>
#> [easypar] 2025-11-21 08:52:11.229506 - Overriding parallel execution setup [FALSE] with global option : FALSE
#>
#>
#> ℹ MOBSTER fits completed in 3.4s.
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 52% [C1] and 48% [C2], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 102, 52%] with mean = 0.58.
#> ● Beta C2 [n = 98, 48%] with mean = 0.11.
#> ℹ Score(s): NLL = -108.12; ICL = -182.58 (-182.58), H = 1.88 (1.88). Fit
#> converged by MM in 70 steps.
# Just 5 resamples of a nonparametric bootstrap run, disabling the parallel engine
options(easypar.parallel = FALSE)
boot_results = mobster_bootstrap(x$best, n.resamples = 5, auto_setup = 'FAST')
#> [ MOBSTER bootstrap ~ 5 resamples from nonparametric bootstrap ]
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 52% [C1] and 48% [C2], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 102, 52%] with mean = 0.58.
#> ● Beta C2 [n = 98, 48%] with mean = 0.11.
#> ℹ Score(s): NLL = -108.12; ICL = -182.58 (-182.58), H = 1.88 (1.88). Fit
#> converged by MM in 70 steps.
#>
#> ℹ Creating nonparametric bootstrap resamples
#> ✔ Creating nonparametric bootstrap resamples ... done
#>
#>
#> ── Running fits ─────────────────────────────────── Might take some time ... ──
#> [easypar] 2025-11-21 08:52:14.716101 - Overriding parallel execution setup [TRUE] with global option : FALSE
#> [ MOBSTER fit ]
#>
#> ✔ Loaded input data, n = 200.
#> ❯ n = 200. Mixture with k = 1,2 Beta(s). Pareto tail: TRUE and FALSE. Output
#> clusters with π > 0.02 and n > 10.
#> ! mobster automatic setup FAST for the analysis.
#> ❯ Scoring (without parallel) 2 x 2 x 2 = 8 models by reICL.
#>
#> [easypar] 2025-11-21 08:52:14.761779 - Overriding parallel execution setup [FALSE] with global option : FALSE
#>
#>
#> ℹ MOBSTER fits completed in 3.8s.
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 50% [C1] and 50% [C2], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 98, 50%] with mean = 0.59.
#> ● Beta C2 [n = 102, 50%] with mean = 0.11.
#> ℹ Score(s): NLL = -101.04; ICL = -168.07 (-168.07), H = 2.23 (2.23). Fit
#> interrupted by MM in 100 steps.
#> [ MOBSTER fit ]
#>
#> ✔ Loaded input data, n = 200.
#> ❯ n = 200. Mixture with k = 1,2 Beta(s). Pareto tail: TRUE and FALSE. Output
#> clusters with π > 0.02 and n > 10.
#> ! mobster automatic setup FAST for the analysis.
#> ❯ Scoring (without parallel) 2 x 2 x 2 = 8 models by reICL.
#>
#> [easypar] 2025-11-21 08:52:18.668438 - Overriding parallel execution setup [FALSE] with global option : FALSE
#>
#>
#> ℹ MOBSTER fits completed in 3.3s.
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 51% [C1] and 49% [C2], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 101, 51%] with mean = 0.59.
#> ● Beta C2 [n = 99, 49%] with mean = 0.11.
#> ℹ Score(s): NLL = -115.21; ICL = -197.42 (-197.42), H = 1.21 (1.21). Fit
#> converged by MM in 62 steps.
#> [ MOBSTER fit ]
#>
#> ✔ Loaded input data, n = 200.
#> ❯ n = 200. Mixture with k = 1,2 Beta(s). Pareto tail: TRUE and FALSE. Output
#> clusters with π > 0.02 and n > 10.
#> ! mobster automatic setup FAST for the analysis.
#> ❯ Scoring (without parallel) 2 x 2 x 2 = 8 models by reICL.
#>
#> [easypar] 2025-11-21 08:52:22.005144 - Overriding parallel execution setup [FALSE] with global option : FALSE
#>
#>
#> ℹ MOBSTER fits completed in 3.3s.
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 50% [C2] and 50% [C1], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 99, 50%] with mean = 0.59.
#> ● Beta C2 [n = 101, 50%] with mean = 0.12.
#> ℹ Score(s): NLL = -108.28; ICL = -182.9 (-182.9), H = 1.87 (1.87). Fit
#> converged by MM in 70 steps.
#> [ MOBSTER fit ]
#>
#> ✔ Loaded input data, n = 200.
#> ❯ n = 200. Mixture with k = 1,2 Beta(s). Pareto tail: TRUE and FALSE. Output
#> clusters with π > 0.02 and n > 10.
#> ! mobster automatic setup FAST for the analysis.
#> ❯ Scoring (without parallel) 2 x 2 x 2 = 8 models by reICL.
#>
#> [easypar] 2025-11-21 08:52:25.44167 - Overriding parallel execution setup [FALSE] with global option : FALSE
#>
#>
#> ℹ MOBSTER fits completed in 3.6s.
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 57% [C1] and 43% [C2], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 113, 57%] with mean = 0.59.
#> ● Beta C2 [n = 87, 43%] with mean = 0.11.
#> ℹ Score(s): NLL = -115.77; ICL = -199.22 (-199.22), H = 0.52 (0.52). Fit
#> converged by MM in 25 steps.
#> [ MOBSTER fit ]
#>
#> ✔ Loaded input data, n = 200.
#> ❯ n = 200. Mixture with k = 1,2 Beta(s). Pareto tail: TRUE and FALSE. Output
#> clusters with π > 0.02 and n > 10.
#> ! mobster automatic setup FAST for the analysis.
#> ❯ Scoring (without parallel) 2 x 2 x 2 = 8 models by reICL.
#>
#> [easypar] 2025-11-21 08:52:29.10236 - Overriding parallel execution setup [FALSE] with global option : FALSE
#>
#>
#> ℹ MOBSTER fits completed in 3.7s.
#>
#> ── [ MOBSTER ] My MOBSTER model n = 200 with k = 2 Beta(s) without tail ────────
#> ● Clusters: π = 55% [C1] and 45% [C2], with π > 0.
#> ✖ No tail fit.
#>
#> ● Beta C1 [n = 109, 55%] with mean = 0.59.
#> ● Beta C2 [n = 91, 45%] with mean = 0.13.
#> ℹ Score(s): NLL = -102.08; ICL = -171.04 (-171.04), H = 1.34 (1.34). Fit
#> converged by MM in 29 steps.
boot_stats = bootstrapped_statistics(x$best, boot_results)
#>
#> ── Computing model frequency ───────────────────────────────────────────────────
#> # A tibble: 1 × 3
#> Model Frequency fit.model
#> <fct> <dbl> <lgl>
#> 1 K = 2 without tail 1 TRUE
#>
#> ── Computing confidence Intervals (CI) for empirical quantiles ─────────────────
#>
#> Mixing proportions
#> # A tibble: 3 × 8
#> cluster statistics min lower_quantile higher_quantile max fit.value
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 C1 Mixing proportion 0.496 0.497 0.563 0.565 0.517
#> 2 C2 Mixing proportion 0.435 0.437 0.503 0.504 0.483
#> 3 Tail Mixing proportion 0 0 0 0 0
#> # ℹ 1 more variable: init.value <dbl>
#>
#> Tail shape/ scale
#> # A tibble: 0 × 8
#> # ℹ 8 variables: cluster <chr>, statistics <chr>, min <dbl>,
#> # lower_quantile <dbl>, higher_quantile <dbl>, max <dbl>, fit.value <dbl>,
#> # init.value <dbl>
#>
#> Beta peaks
#> # A tibble: 4 × 8
#> cluster statistics min lower_quantile higher_quantile max fit.value
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 C1 Mean 0.586 0.586 0.592 0.593 0.581
#> 2 C1 Variance 0.00682 0.00690 0.00880 0.00885 0.00893
#> 3 C2 Mean 0.110 0.110 0.128 0.129 0.112
#> 4 C2 Variance 0.00301 0.00309 0.00446 0.00449 0.00341
#> # ℹ 1 more variable: init.value <dbl>
#>
#> ── Computing co-clustering probability for nonparametric bootstrap ─────────────
plot_bootstrap_coclustering(x$best, boot_results, boot_stats)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's colour values.