Determine the optimal error tolerance to QC clonal simple CNAs.
Source:R/auto_tolerance.R
auto_tolerance.RdThe QC procedure implemented by function analyze_peaks allows to
pass or fail a sample based on a custom purity error (maximum error to allow).
A regression has been used to measure the rate of false positives
from simulated tumours with variable coverage and purity. This allows to determine
an optimal value of $\(\epsilon\)$, the parameter `purity_error` of function
analyze_peaks, for a desired rate of alse positives basde on the sample
coverage and putative purity.
auto_tolerance(purity, coverage, fpr = 0.1, epsilon_range = c(0.01, 0.08))Arguments
Value
The $\(\epsilon\)$ value estimated from data, constrained to be in `epsilon_range`, in order to limit the false positive rate to be at most `fpr`.
Examples
# Desired 10% FPR for a 30% pure tumour at 90x
auto_tolerance(.3, 90)
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance above 100% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance above 100% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance above 100% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance above 100% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance above 100% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance below 1% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `epsilon = inv_fun(fpr)`.
#> Caused by warning in `inv_fun()`:
#> ! Tolerance above 100% for FPR = 0.1 - returning 0.01 (epsilon_range).
#> → Inverting training from regression; requiring epsilon in range [0.01 - 0.08].
#> → Generating interpolated 2D regression map.
#> Warning: There were 15352 warnings in `mutate()`.
#> The first warning was:
#> ℹ In argument: `fit = `%>%`(...)`.
#> ℹ In row 1.
#> Caused by warning in `akima::interp()`:
#> ! collinear points, trying to add some jitter to avoid colinearities!
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 15351 remaining warnings.
#> Warning: collinear points, trying to add some jitter to avoid colinearities!
#> Warning: success: collinearities reduced through jitter
#> ✔ Suggested point estimate: ε = 0.0121101139946641.
#> $epsilon_range
#> [1] 0.01211011
#>
#> $training_plot
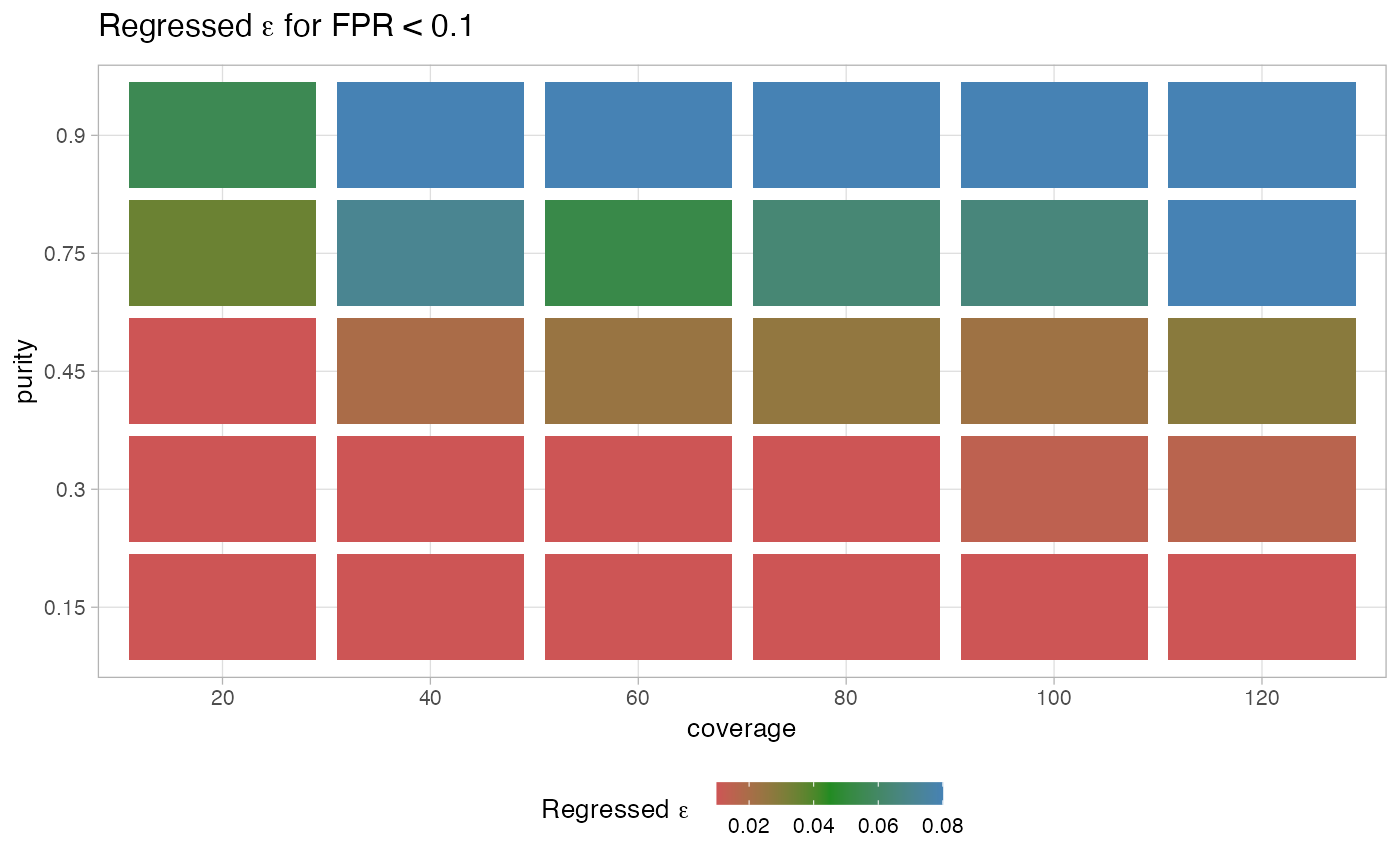 #>
#> $extrapolation_plot
#>
#> $extrapolation_plot
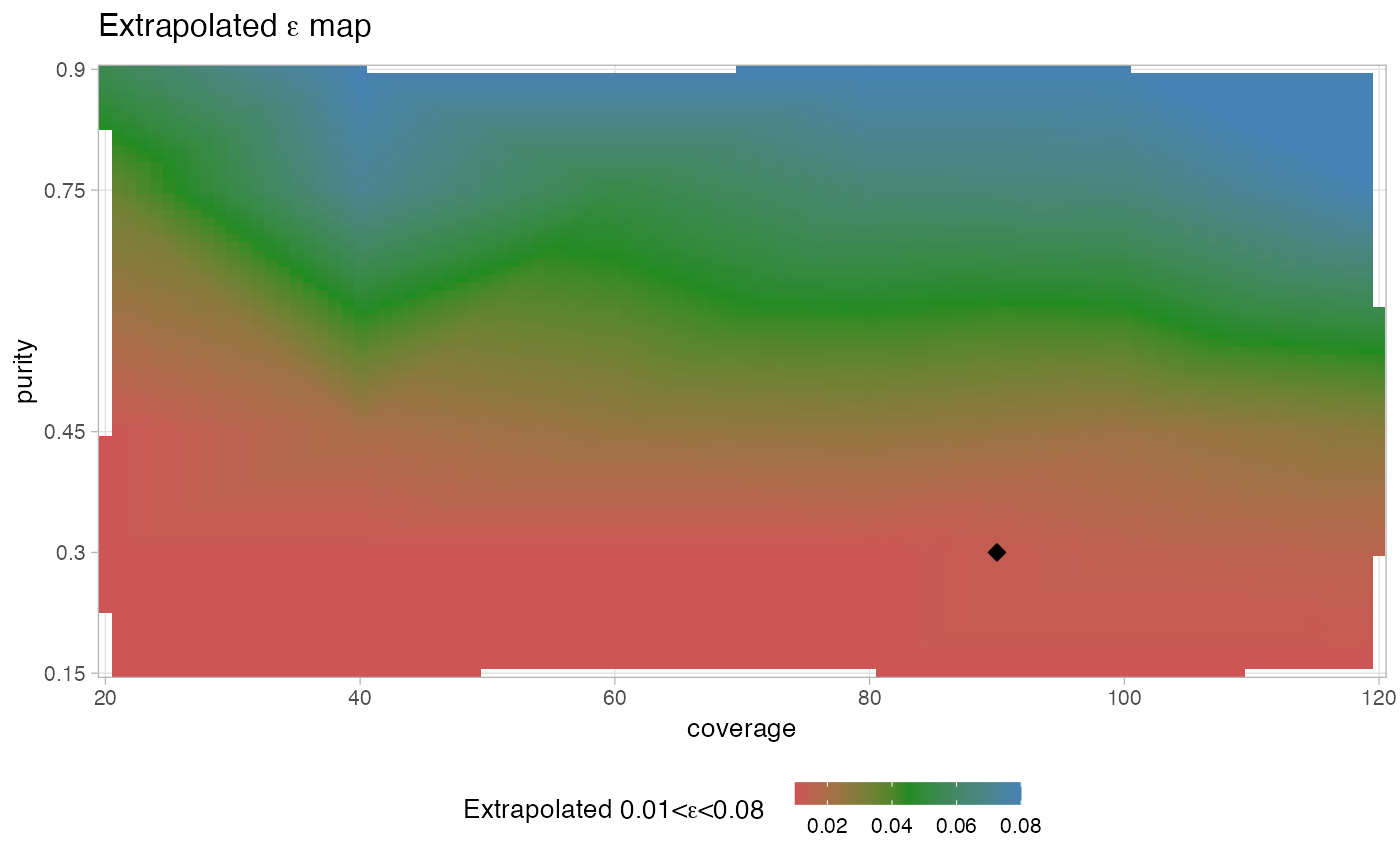 #>
#>