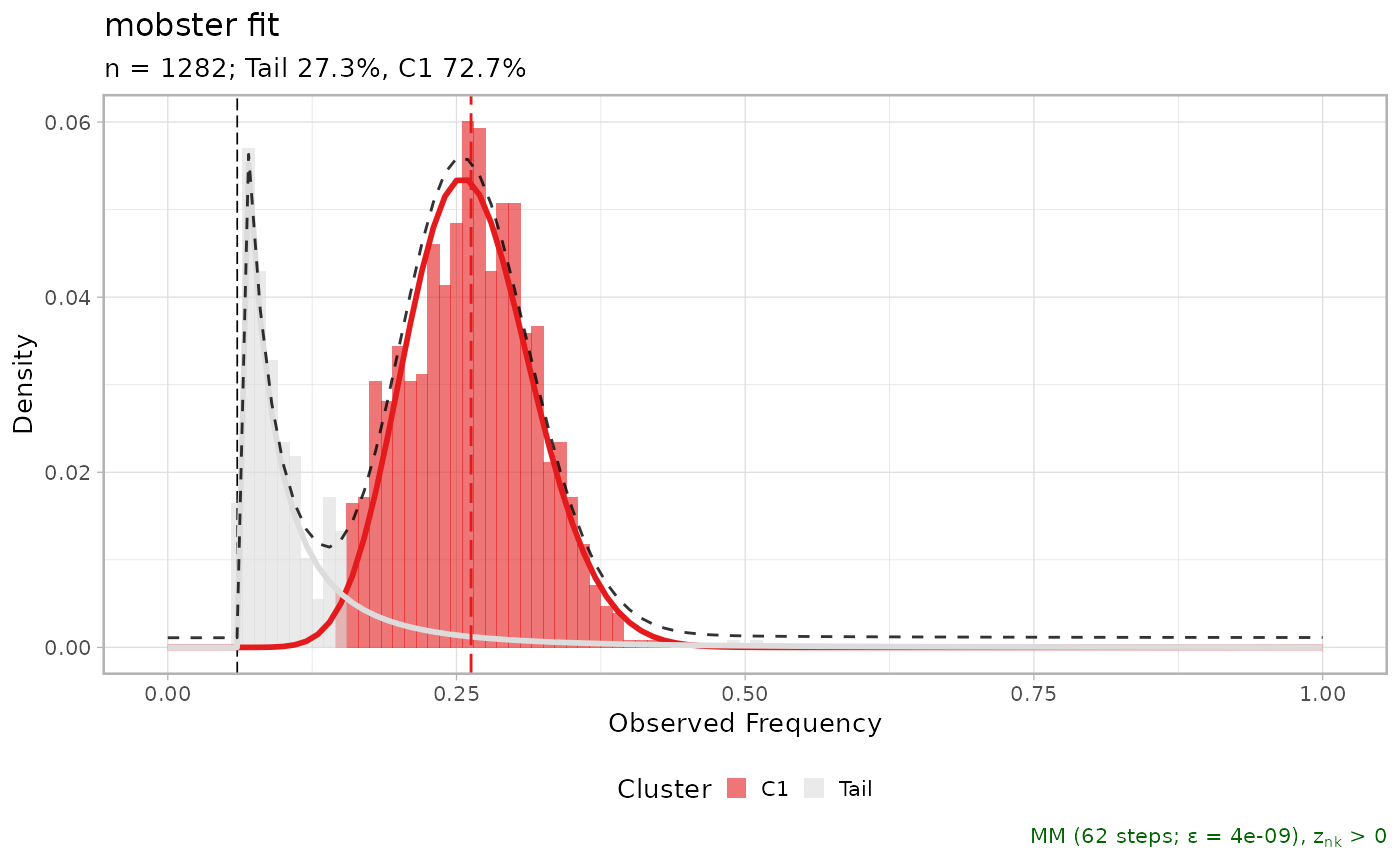
MOBSTER fit for the LU4 lung sample; this object is the result of running `mobster_fit` function on the input data described in the main MOBSTER paper. The data consists of diploid mutations for the sample available at the Comprehensive Omics Archive of Lung Adenocarcinoma (http://genome.kaist.ac.kr/).
Usage
data(LU4_lung_sample)Examples
data(LU4_lung_sample, package = 'mobster')
print(LU4_lung_sample$best)
#> ── [ MOBSTER ] n = 1282 with k = 1 Beta(s) and a tail ─────────────────────────
#> ● Clusters: π = 73% [C1] and 27% [Tail], with π > 0.
#> ● Tail [n = 310, 27%] with alpha = 1.9.
#> ● Beta C1 [n = 972, 73%] with mean = 0.26.
#> ℹ Score(s): NLL = -1527.89; ICL = -2797.42 (-3012.83), H = 215.41 (0). Fit
#> converged by MM in 62 steps.
plot(LU4_lung_sample$best)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the mobster package.
#> Please report the issue at <https://github.com/caravagnalab/mobster/issues>.
#> Warning: The `<scale>` argument of `guides()` cannot be `FALSE`. Use "none" instead as
#> of ggplot2 3.3.4.
#> ℹ The deprecated feature was likely used in the mobster package.
#> Please report the issue at <https://github.com/caravagnalab/mobster/issues>.
#> Warning: The dot-dot notation (`..count..`) was deprecated in ggplot2 3.4.0.
#> ℹ Please use `after_stat(count)` instead.
#> ℹ The deprecated feature was likely used in the mobster package.
#> Please report the issue at <https://github.com/caravagnalab/mobster/issues>.