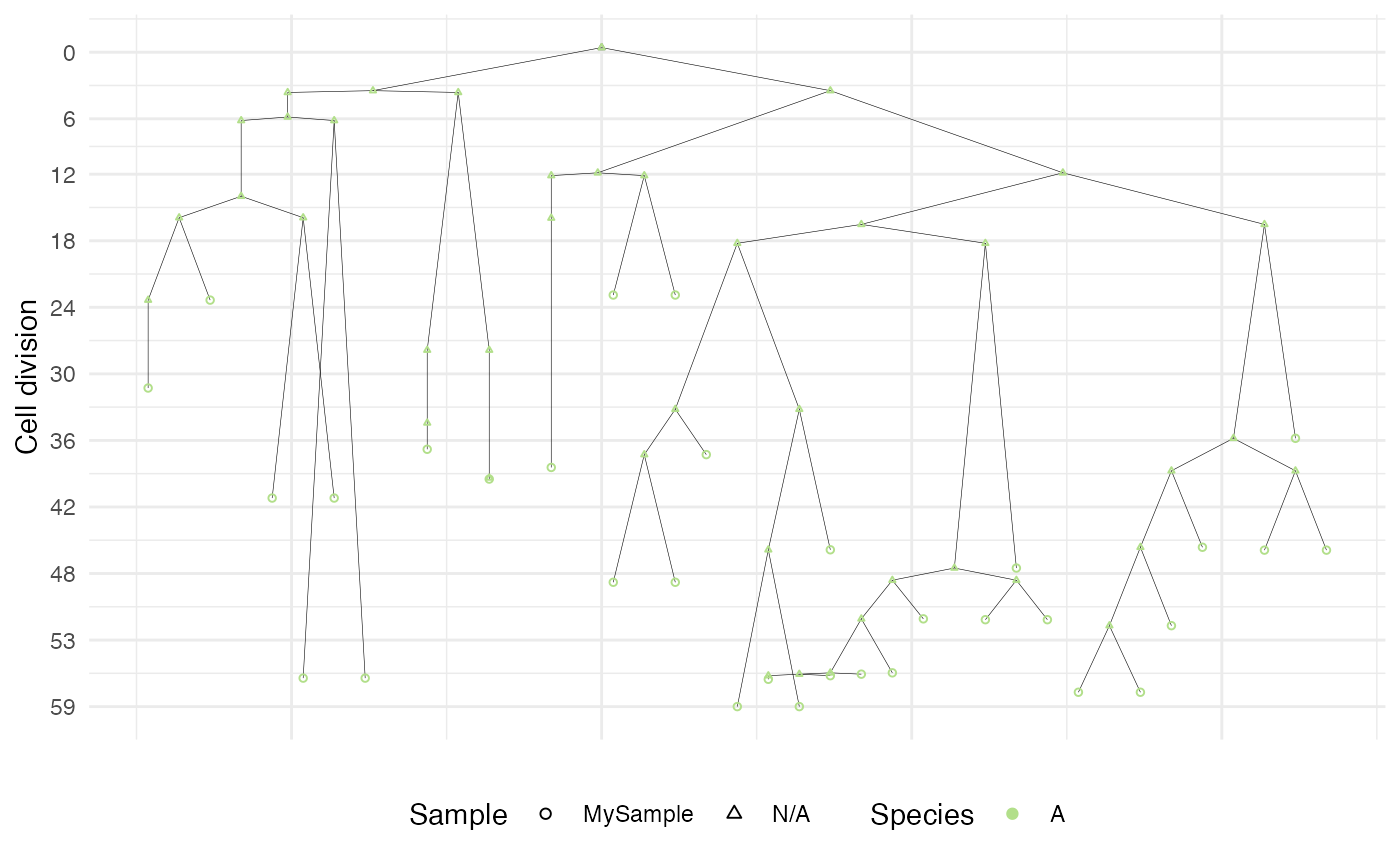
Plot the samples cell division forest. This plot is carried out using
ggraph and for simplicity of visualisation the forest is plot as a
set of trees connected to a generic wildtype cell.
Examples
sim <- SpatialSimulation()
sim$add_mutant(name = "A", growth_rates = 0.08, death_rates = 0.0)
sim$place_cell("A", 500, 500)
sim$run_up_to_time(60)
#>
[████████████████████████████████████████] 100% [00m:00s] Saving snapshot
sim$sample_cells("MySample", c(500, 500), c(510, 510))
forest <- sim$get_samples_forest()
plot_forest(forest)
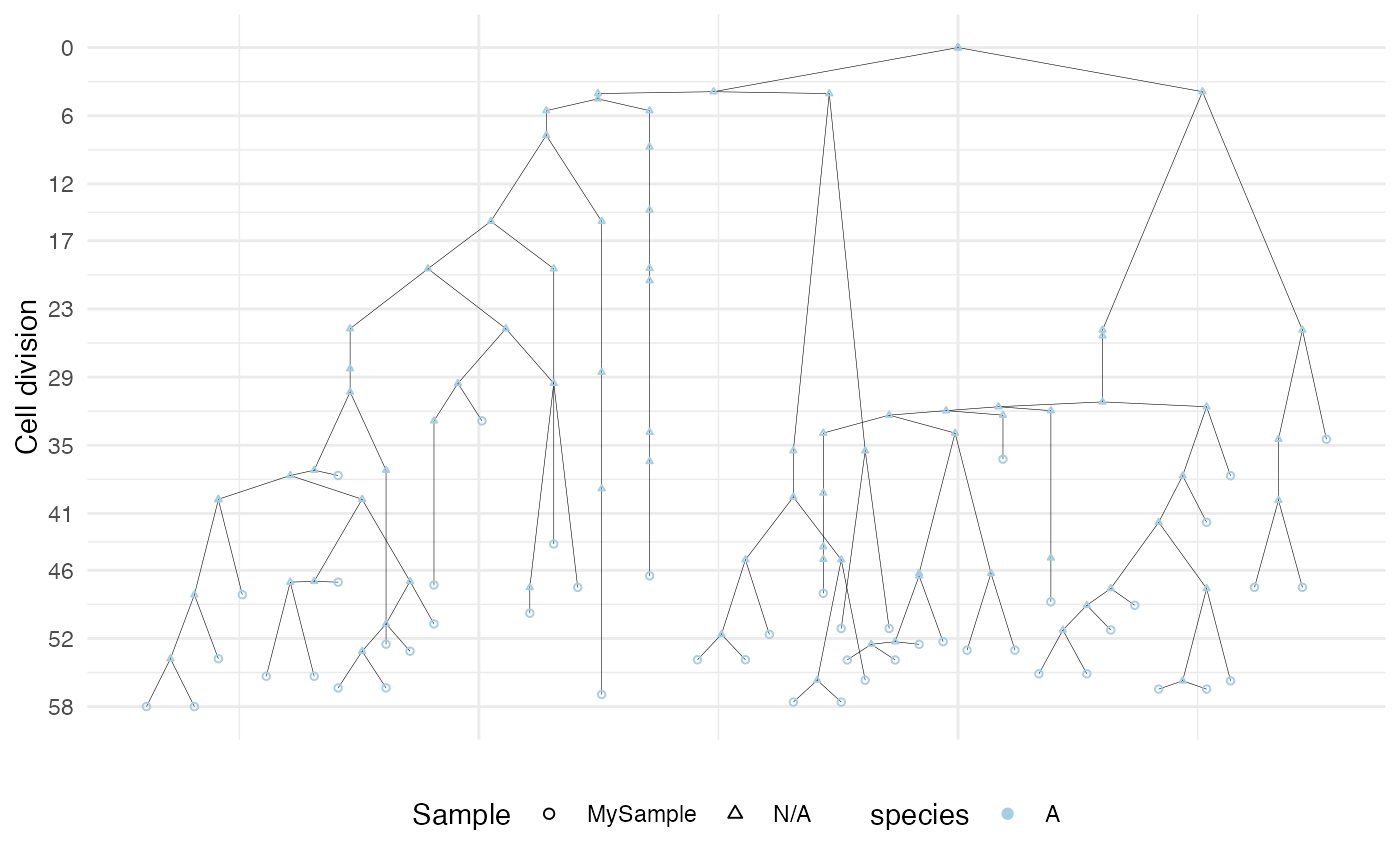 # define a custom color map
color_map <- c("#B2DF8A")
names(color_map) <- c("A")
plot_forest(forest, color_map=color_map)
# define a custom color map
color_map <- c("#B2DF8A")
names(color_map) <- c("A")
plot_forest(forest, color_map=color_map)