rRACES wraps the RACES simulation and data-generation engine by using the R/C++ Rcpp interface, making it easy to “program” stochastic tumour evolution.
Pipeline overview

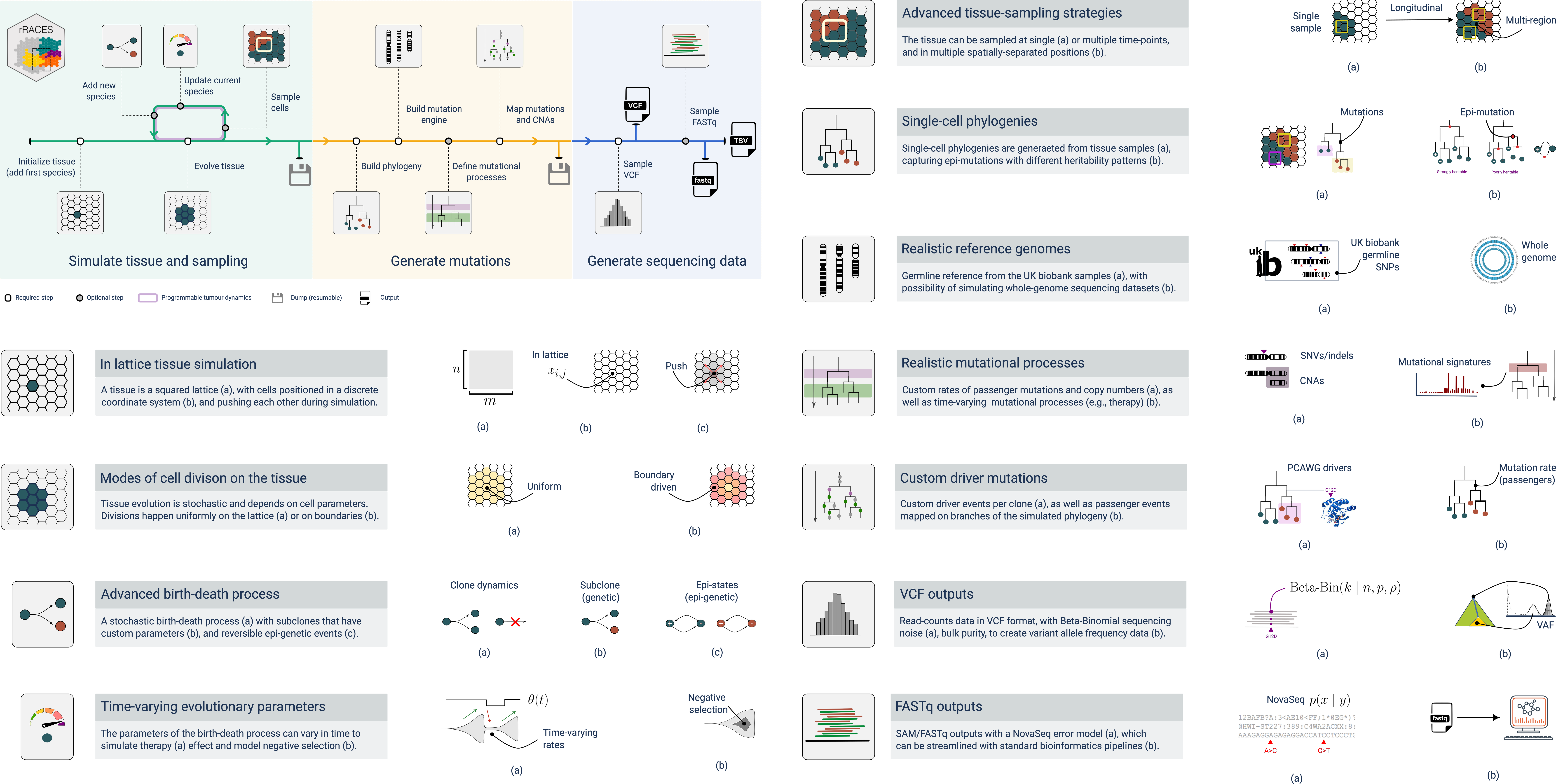
The high-level overview of pipeline of rRACES is a three-steps process, where:
a discrete tissue is first simulated, with distinct clones defined by stochastic rates of growth and death. These compete to colonise the tissue in a standard stochastic birth-death process, which in rRACES is empowered by a time-varying structure that allows weasily modelling of complex evolutionary forces that vary over time (e.g., therapy-induced negative selection). The tissue can be sampled at multiple time-points and in multiple spatially-separated positions.
from the sampled cells, the simulated phylogenetic history of the tumour can be extracted. The distance among the involved cells will represent the lineage history of the simulated clones, and mutational processes can be used to attach genomes to each simulated cells. rRACES supports standard germline and somatic mutations, including single-nucleotide variants (SNVs), insertion-deletions (indels) and copy number alterations (CNAs). For SNVs and indels time-varying mutational signatures can also be included.
from the simulated phylogenetic history of the tumour it is possible to generate tumour-matched-normal synthetic data for benchmarking bioinformatics tools, or to infer parameters against real sequencing data. rRACES output include common VCF formats, as well as low-level SAM outputs that can be used for aligment and downstream analysis. Every simulated sample can be assigned custom coverage and purity.
Main features
The rRACES engine has a number of features, the most relevant are:
- cells are associated with species that proliferate in a stochastic fashion on a 2D lattice;
- every species is defined by (i) an abstract “mutant” and (ii) a epi-state; the epi-state is binary (0/1, on-off). The combination of the mutant and the epi-state determine the evolutionary parameters of each species, which can can change over time mimicking treatment-related evolutionary pressures, for instance.
- Species evolution is stochastic and follows a no-back mutations model, where at every cell division the mutant status heritable, while the epistate can be stochastically reversible;
- at the molecular level, the genome of tumour cells are mutated by simple mutations (SNVs and indels) and more complex CNAs. Mutations - in the broad sense - can be either passengers or drivers ones, and can be linked to realistic mutational processes. All their rates of accumulation can change over time, mimicking the emergence of time-varying mutational forces (e.g., to model treatment). Moreover, realistic germline samples can be simulated by interfacing with the UK biobank database.
- arbitrary tissue sampling schema can be simulated, including multi-region and longitudinal datasets with any number of samples and time-points;
- realistic read-counts based bulk sequencing data can be generated, both at the level of whole-genome, whole-exome and targeted panels, as well as at the level of pre-processed outputs (VCFs) or low-level sequencing outputs (SAM);
- full access to the evolutionary process and its output is available, allowing easy testing of complex clonal architecture identification methods, mutation callers, etc.
Detailed pipeline
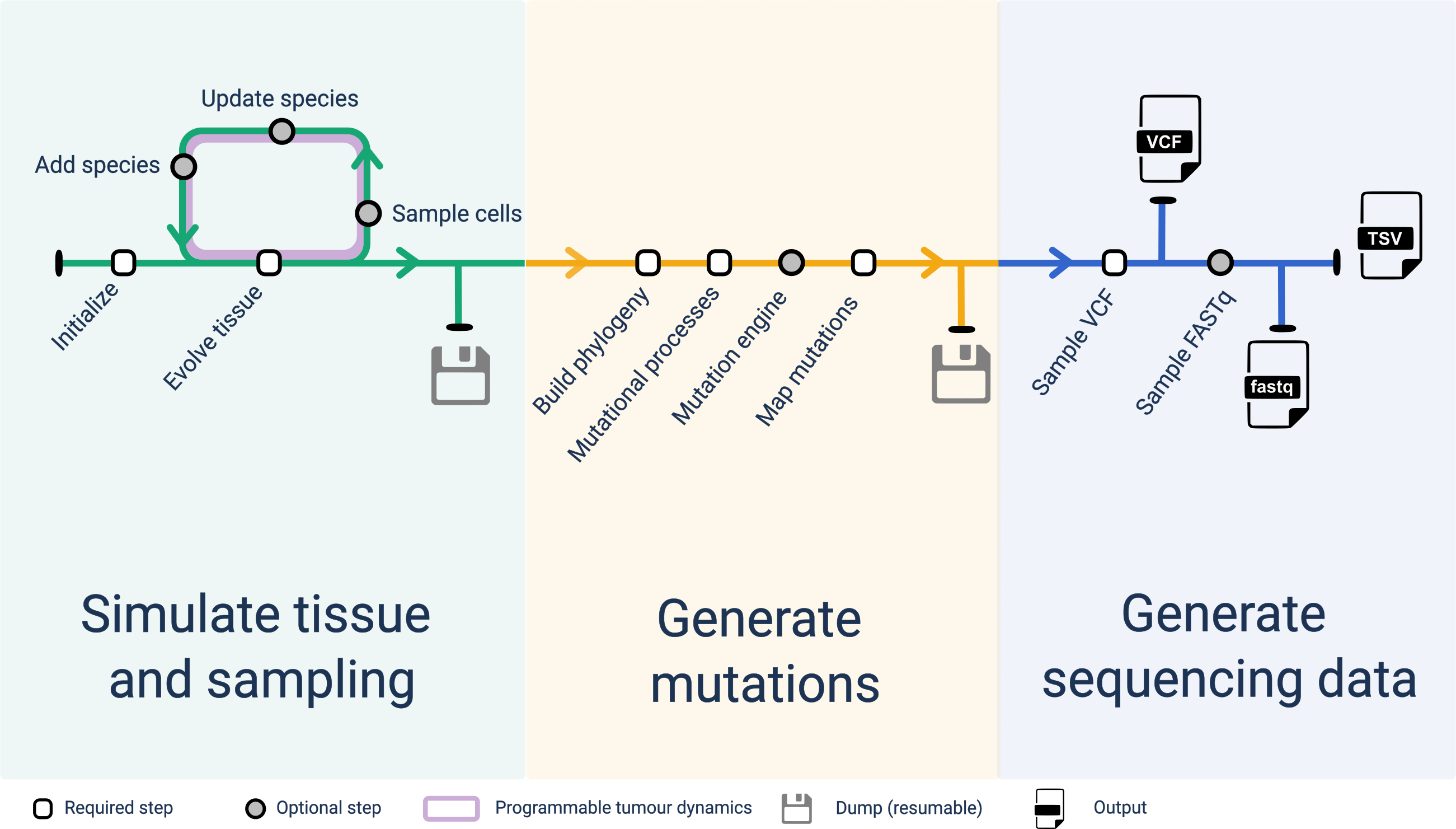
A rRACES simulation follows the steps (some required, some optional) that we disucssd below. The state of the simulation can be saved (and resumed) at several time-points.
Tissue simulation
- Initialise: a squared tissue is initialised, a species is defined and a single cell placed onto it;
- Evolve tissue: all the cells on the tissue are left to grow stochastically;
- Sample cells: cells can be sampled from the tissue to mimic a measurement;
- Updates/add species: the parameters of each species can be modified, and cells from new species can be defined and added to the tissue (subclones).
Steps 2-4 represent an iterative interface for “programmable tumour dynamics” that gives the user the flexibility to code a custom evolutionary process.
Mutations generation
- Phylogeny: from sampled cells, a phylogeny is built that reflects the evolutionary history of the simulated tumour.
- Mutational processes: mutational processes can be mapped onto the temporal-evolution of the process.
- Mutational engine: an engine to simulate mutations is built using a real reference genome.
- Map mutations: mutations are stochatically attached to the simulated phylogeny.
Sequencing data generation
- VCF: given mutations on the phylogenetic tree, custom coverage and sample purity, a VCF can be simulated to mimic variant-calling with Beta-Binomial noise.
- SAM: from the same mutations it is possible to generate reads for a tumour matched-normal assay, and fed that data to a custom biorinformatics pipeline.