Plots the signatures exposure changes along a phylogenetic forest.
Examples
sim <- SpatialSimulation()
sim$add_mutant(name = "A",
growth_rates = 0.2,
death_rates = 0.0)
sim$place_cell("A", 500, 500)
sim$run_up_to_time(150)
#>
[████████████████████████████████████████] 100% [00m:00s] Saving snapshot
# sampling tissue
n_w <- n_h <- 50
ncells <- 0.8 * n_w * n_h
bbox <- sim$search_sample(c("A" = ncells), n_w, n_h)
sim$sample_cells("Sampling", bbox$lower_corner, bbox$upper_corner)
forest <- sim$get_samples_forest()
# placing mutations
m_engine <- MutationEngine(setup_code = "demo")
#> Downloading reference genome...
#> Reference genome downloaded
#> Decompressing reference file...done
#> Downloading SBS file...
#> SBS file downloaded
#> Downloading indel file...
#> indel file downloaded
#> Downloading driver mutation file...
#> Driver mutation file downloaded
#> Downloading passenger CNAs file...
#> Passenger CNAs file downloaded
#> Downloading germline mutations...
#> Germline mutations downloaded
#> Building context index...
#>
[█---------------------------------------] 0% [00m:00s] Processing chr. 22
[██████████------------------------------] 24% [00m:01s] Processing chr. 22
[████████████████████--------------------] 49% [00m:02s] Processing chr. 22
[██████████████████████████████----------] 73% [00m:03s] Processing chr. 22
[████████████████████████████████████████] 98% [00m:04s] Processing chr. 22
[████████████████████████████████████████] 100% [00m:04s] Context index built
#>
[█---------------------------------------] 0% [00m:00s] Saving context index
[████████████████████████████████████████] 100% [00m:00s] Context index saved
#> done
#> Building repeated sequence index...
#>
[█---------------------------------------] 0% [00m:00s] Processing chr. 22
[█---------------------------------------] 0% [00m:00s] Processing chr. 22
[█---------------------------------------] 0% [00m:01s] Processing chr. 22
[█---------------------------------------] 0% [00m:05s] Processing chr. 22
[█---------------------------------------] 0% [00m:07s] Processing chr. 22
[█---------------------------------------] 0% [00m:08s] Processing chr. 22
[█---------------------------------------] 0% [00m:10s] Processing chr. 22
[█---------------------------------------] 0% [00m:11s] Processing chr. 22
[█---------------------------------------] 0% [00m:16s] Processing chr. 22
[████████████████████████████████████████] 100% [00m:17s] RS index built
#>
[█---------------------------------------] 0% [00m:00s] Saving RS index
[█---------------------------------------] 0% [00m:00s] Saving RS index
[█---------------------------------------] 0% [00m:01s] Saving RS index
[█████████████---------------------------] 31% [00m:03s] Saving RS index
[██████████████████████████████████------] 83% [00m:04s] Saving RS index
done
#>
[████████████████████████████████████████] 100% [00m:04s] RS index saved
#>
[█---------------------------------------] 0% [00m:00s] Loading germline
[████████████████████████████████████████] 100% [00m:00s] Germline loaded
#>
[█---------------------------------------] 0% [00m:00s] Saving germline
[████████████████████████████████████████] 100% [00m:00s] Germline saved
m_engine$add_mutant(mutant_name = "A",
passenger_rates = c(SNV = 8e-8))
#>
[█---------------------------------------] 0% [00m:00s] Retrieving "A" SNVs
[████████████████████████████████████████] 100% [00m:00s] "A" SNVs retrieved
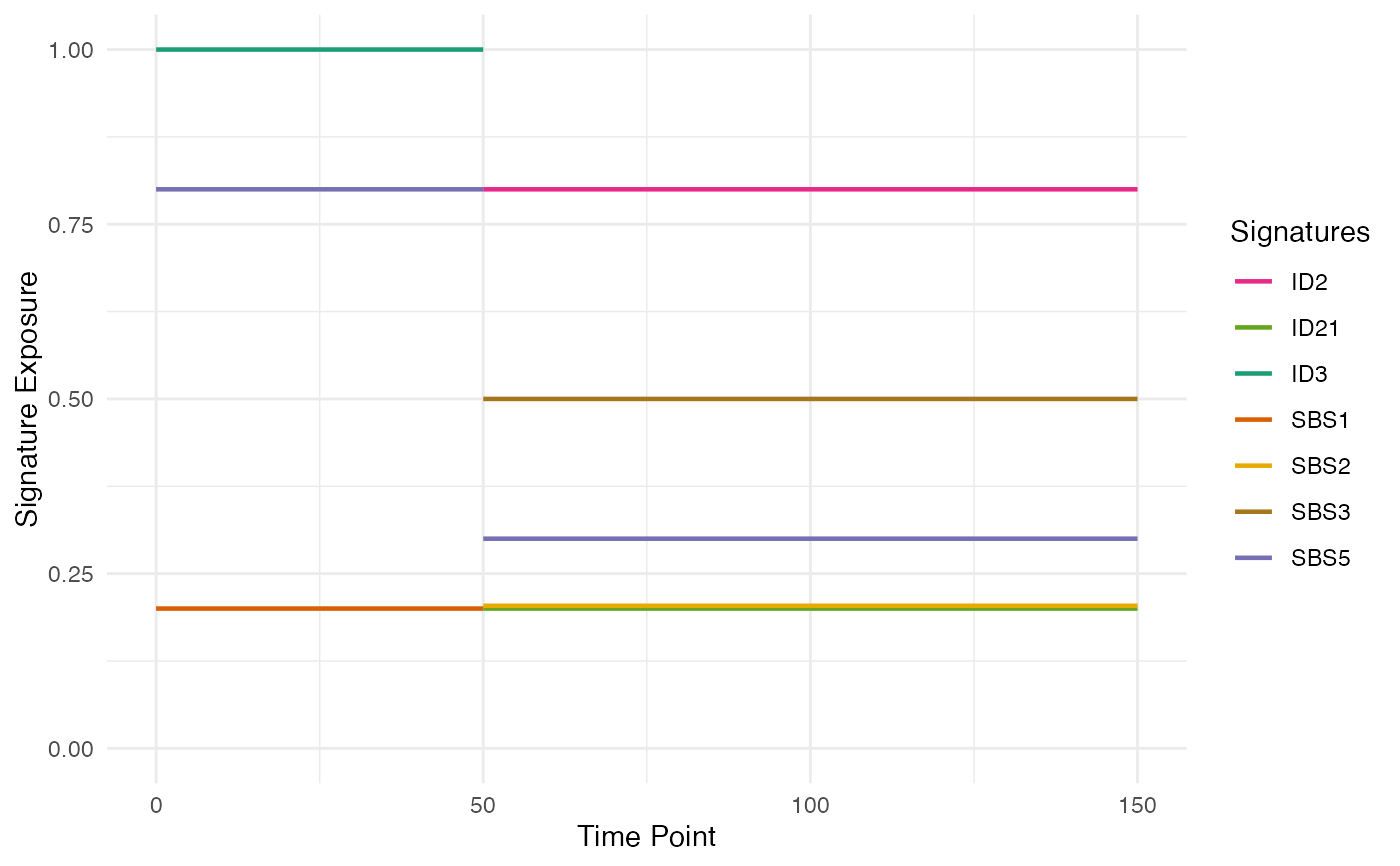
m_engine$add_exposure(c(SBS1 = 0.2, SBS5 = 0.8, ID3 = 1))
m_engine$add_exposure(time = 50,
c(SBS5 = 0.3, SBS2 = 0.2, SBS3 = 0.5,
ID2 = 0.8, ID21 = 0.2))
phylo_forest <- m_engine$place_mutations(forest, 500, 10)
#>
[█---------------------------------------] 0% [00m:00s] Placing mutations
[████████████████████████████████████████] 100% [00m:00s] Mutations placed
# plotting the phylogenetic forest
plot_exposure_timeline(phylo_forest)
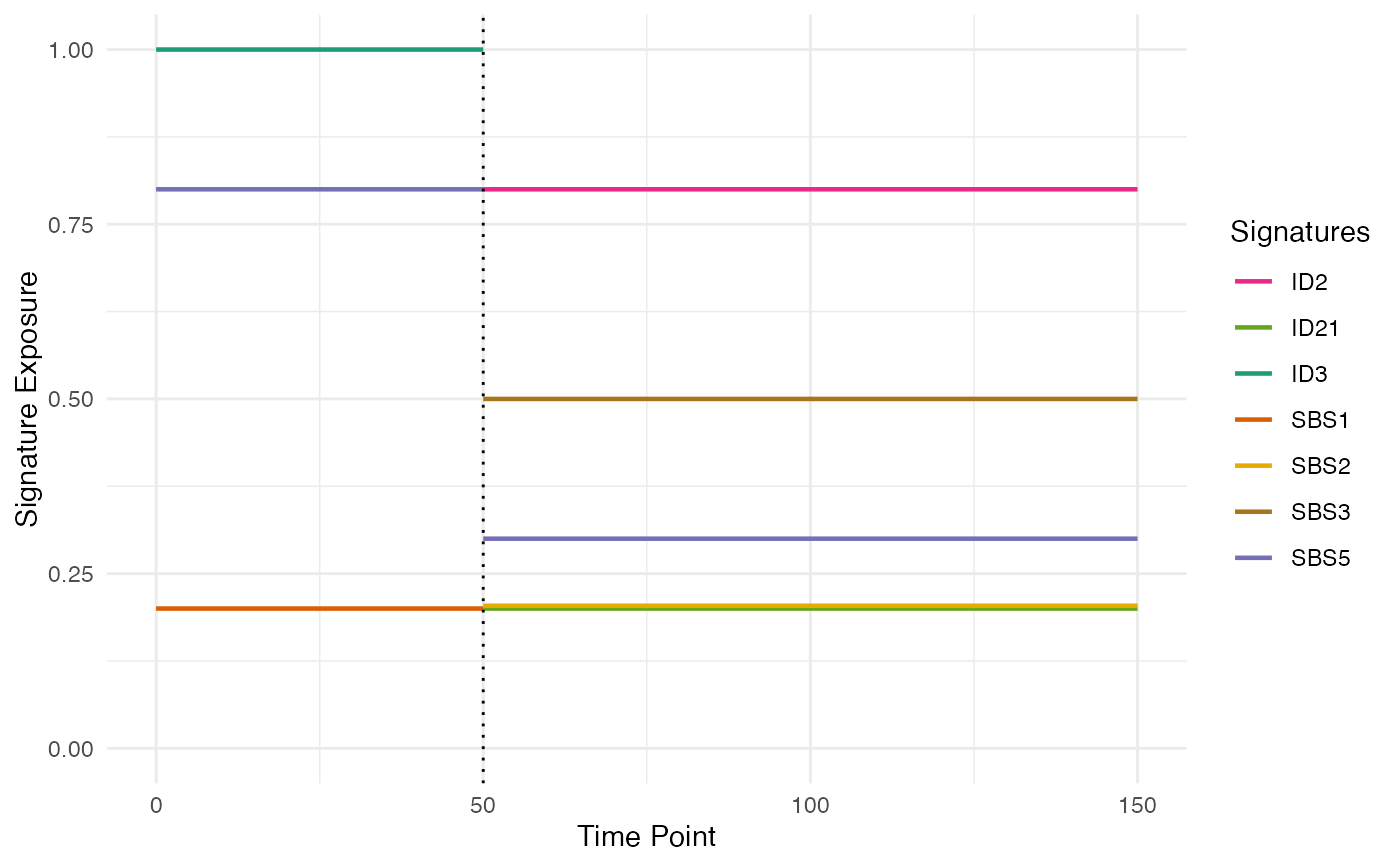
 # plotting the phylogenetic forest emphatizing the exposure switches
plot_exposure_timeline(phylo_forest, emphatize_switches=TRUE)
# plotting the phylogenetic forest emphatizing the exposure switches
plot_exposure_timeline(phylo_forest, emphatize_switches=TRUE)
 # deleting the mutation engine directory
unlink("demo", recursive=TRUE)
# deleting the mutation engine directory
unlink("demo", recursive=TRUE)
