
Plot Histogram of Variant Allele Frequency (VAF)
Source:R/plot_vaf_histogram.R
plot_VAF_histogram.RdThis function generates a histogram showing the distribution of Variant Allele Frequency (VAF) across samples and chromosomes.
Usage
plot_VAF_histogram(
seq_result,
chromosomes = NULL,
samples = NULL,
labels = NULL,
binwidth = NULL,
cuts = c(0, 1)
)Arguments
- seq_result
A data frame containing sequencing results.
- chromosomes
A character vector specifying the chromosomes to include in the plot (default: all the chromosomes in
seq_res).- samples
A character vector specifying the sample names to include in the plot. When set to
NULL, the function includes all samples except the "normal_sample" (default:NULL).- binwidth
The width of the plot bins. When set to
NULL, the function computes the most convinient bin width according to the maximum coverage reported in the dataframe (default:NULL).- cuts
A numeric vector specifying the range of VAF values to include in the plot (default:
c(0, 1)).- colour_by
A character indicating whether to color the histogram bars by "causes" or "classes" (default: "causes").
Examples
# set the seed of the random number generator
set.seed(0)
sim <- SpatialSimulation()
sim$add_mutant(name = "A",
growth_rates = 0.1,
death_rates = 0.0)
sim$place_cell("A", 500, 500)
sim$run_up_to_time(100)
#>
[████████████████████████████████████████] 100% [00m:00s] Saving snapshot
# sampling tissue
n_w <- n_h <- 10
ncells <- 0.8 * n_w * n_h
bbox <- sim$search_sample(c("A" = ncells), n_w, n_h)
sim$sample_cells("SampleA", bbox$lower_corner, bbox$upper_corner)
# adding second mutant
sim$add_mutant(name = "B",
growth_rates = 0.3,
death_rates = 0.0)
sim$mutate_progeny(sim$choose_cell_in("A"), "B")
sim$run_up_to_time(300)
#>
[████████████████████████████████████████] 100% [00m:00s] Saving snapshot
# sampling tissue again
bbox <- sim$search_sample(c("B" = ncells), n_w, n_h)
sim$sample_cells("SampleB", bbox$lower_corner, bbox$upper_corner)
forest <- sim$get_samples_forest()
# placing mutations
m_engine <- MutationEngine(setup_code = "demo")
#> Downloading reference genome...
#> Reference genome downloaded
#> Decompressing reference file...done
#> Downloading SBS file...
#> SBS file downloaded
#> Downloading indel file...
#> indel file downloaded
#> Downloading driver mutation file...
#> Driver mutation file downloaded
#> Downloading passenger CNAs file...
#> Passenger CNAs file downloaded
#> Downloading germline mutations...
#> Germline mutations downloaded
#> Building context index...
#>
[█---------------------------------------] 0% [00m:00s] Processing chr. 22
[██████████------------------------------] 24% [00m:01s] Processing chr. 22
[████████████████████--------------------] 49% [00m:02s] Processing chr. 22
[██████████████████████████████----------] 73% [00m:03s] Processing chr. 22
[████████████████████████████████████████] 98% [00m:04s] Processing chr. 22
[████████████████████████████████████████] 100% [00m:04s] Context index built
#>
[█---------------------------------------] 0% [00m:00s] Saving context index
[████████████████████████████████████████] 100% [00m:00s] Context index saved
#> done
#> Building repeated sequence index...
#>
[█---------------------------------------] 0% [00m:00s] Processing chr. 22
[█---------------------------------------] 0% [00m:00s] Processing chr. 22
[█---------------------------------------] 0% [00m:01s] Processing chr. 22
[█---------------------------------------] 0% [00m:05s] Processing chr. 22
[█---------------------------------------] 0% [00m:07s] Processing chr. 22
[█---------------------------------------] 0% [00m:09s] Processing chr. 22
[█---------------------------------------] 0% [00m:11s] Processing chr. 22
[█---------------------------------------] 0% [00m:13s] Processing chr. 22
[█---------------------------------------] 0% [00m:18s] Processing chr. 22
[████████████████████████████████████████] 100% [00m:18s] RS index built
#>
[█---------------------------------------] 0% [00m:00s] Saving RS index
[█---------------------------------------] 0% [00m:00s] Saving RS index
[█---------------------------------------] 2% [00m:02s] Saving RS index
[████████████████████████----------------] 58% [00m:03s] Saving RS index
done
#>
[████████████████████████████████████████] 100% [00m:03s] RS index saved
#>
[█---------------------------------------] 0% [00m:00s] Loading germline
[████████████████████████████████████████] 100% [00m:00s] Germline loaded
#>
[█---------------------------------------] 0% [00m:00s] Saving germline
[████████████████████████████████████████] 100% [00m:00s] Germline saved
m_engine$add_mutant(mutant_name="A", passenger_rates=c(SNV=5e-8))
#>
[█---------------------------------------] 0% [00m:00s] Retrieving "A" SNVs
[████████████████████████████████████████] 100% [00m:00s] "A" SNVs retrieved
m_engine$add_mutant(mutant_name="B", passenger_rates=c(SNV=5e-9))
#>
[█---------------------------------------] 0% [00m:00s] Retrieving "B" SNVs
[████████████████████████████████████████] 100% [00m:00s] "B" SNVs retrieved
m_engine$add_exposure(c(SBS1 = 0.2, SBS5 = 0.8))
phylo_forest <- m_engine$place_mutations(forest, 10, 10)
#>
[█---------------------------------------] 0% [00m:00s] Placing mutations
[████████████████████████████████████████] 100% [00m:00s] Mutations placed
# simulating sequencing without the normal sample
seq_results <- simulate_seq(phylo_forest, coverage = 10, write_SAM = F,
with_normal_sample = FALSE)
#>
[█---------------------------------------] 0% [00m:00s] Found 22
[█---------------------------------------] 0% [00m:00s] Reading 22
[█---------------------------------------] 0% [00m:01s] Reading 22
[█---------------------------------------] 0% [00m:02s] Reading 22
[█---------------------------------------] 0% [00m:02s] Processing chr. 22
[█---------------------------------------] 0% [00m:03s] Processing chr. 22
[█---------------------------------------] 1% [00m:04s] Processing chr. 22
[█---------------------------------------] 2% [00m:05s] Processing chr. 22
[██--------------------------------------] 3% [00m:06s] Processing chr. 22
[██--------------------------------------] 4% [00m:07s] Processing chr. 22
[███-------------------------------------] 5% [00m:08s] Processing chr. 22
[███-------------------------------------] 6% [00m:09s] Processing chr. 22
[███-------------------------------------] 7% [00m:10s] Processing chr. 22
[████------------------------------------] 8% [00m:11s] Processing chr. 22
[████------------------------------------] 9% [00m:12s] Processing chr. 22
[█████-----------------------------------] 10% [00m:13s] Processing chr. 22
[█████-----------------------------------] 11% [00m:14s] Processing chr. 22
[█████-----------------------------------] 12% [00m:15s] Processing chr. 22
[██████----------------------------------] 14% [00m:16s] Processing chr. 22
[███████---------------------------------] 15% [00m:17s] Processing chr. 22
[███████---------------------------------] 16% [00m:18s] Processing chr. 22
[███████---------------------------------] 17% [00m:19s] Processing chr. 22
[████████--------------------------------] 18% [00m:20s] Processing chr. 22
[████████--------------------------------] 19% [00m:21s] Processing chr. 22
[█████████-------------------------------] 20% [00m:22s] Processing chr. 22
[█████████-------------------------------] 21% [00m:23s] Processing chr. 22
[█████████-------------------------------] 22% [00m:24s] Processing chr. 22
[██████████------------------------------] 23% [00m:25s] Processing chr. 22
[██████████------------------------------] 24% [00m:26s] Processing chr. 22
[███████████-----------------------------] 25% [00m:27s] Processing chr. 22
[███████████-----------------------------] 26% [00m:28s] Processing chr. 22
[███████████-----------------------------] 27% [00m:29s] Processing chr. 22
[████████████----------------------------] 29% [00m:30s] Processing chr. 22
[█████████████---------------------------] 30% [00m:31s] Processing chr. 22
[█████████████---------------------------] 31% [00m:32s] Processing chr. 22
[█████████████---------------------------] 32% [00m:33s] Processing chr. 22
[██████████████--------------------------] 33% [00m:34s] Processing chr. 22
[██████████████--------------------------] 34% [00m:35s] Processing chr. 22
[███████████████-------------------------] 35% [00m:36s] Processing chr. 22
[███████████████-------------------------] 36% [00m:37s] Processing chr. 22
[███████████████-------------------------] 37% [00m:38s] Processing chr. 22
[████████████████------------------------] 38% [00m:39s] Processing chr. 22
[████████████████------------------------] 39% [00m:40s] Processing chr. 22
[█████████████████-----------------------] 40% [00m:41s] Processing chr. 22
[█████████████████-----------------------] 41% [00m:42s] Processing chr. 22
[██████████████████----------------------] 43% [00m:43s] Processing chr. 22
[██████████████████----------------------] 44% [00m:44s] Processing chr. 22
[███████████████████---------------------] 45% [00m:45s] Processing chr. 22
[███████████████████---------------------] 46% [00m:46s] Processing chr. 22
[███████████████████---------------------] 47% [00m:47s] Processing chr. 22
[████████████████████--------------------] 48% [00m:48s] Processing chr. 22
[████████████████████--------------------] 49% [00m:49s] Processing chr. 22
[█████████████████████-------------------] 50% [00m:50s] Processing chr. 22
[█████████████████████-------------------] 51% [00m:51s] Processing chr. 22
[█████████████████████-------------------] 52% [00m:52s] Processing chr. 22
[██████████████████████------------------] 52% [00m:53s] Processing chr. 22
[██████████████████████------------------] 54% [00m:54s] Processing chr. 22
[███████████████████████-----------------] 55% [00m:55s] Processing chr. 22
[███████████████████████-----------------] 55% [00m:56s] Processing chr. 22
[███████████████████████-----------------] 56% [00m:57s] Processing chr. 22
[███████████████████████-----------------] 57% [00m:58s] Processing chr. 22
[████████████████████████----------------] 58% [00m:59s] Processing chr. 22
[████████████████████████----------------] 58% [01m:00s] Processing chr. 22
[█████████████████████████---------------] 61% [01m:01s] Processing chr. 22
[█████████████████████████---------------] 61% [01m:02s] Processing chr. 22
[█████████████████████████---------------] 62% [01m:03s] Processing chr. 22
[██████████████████████████--------------] 64% [01m:04s] Processing chr. 22
[███████████████████████████-------------] 65% [01m:05s] Processing chr. 22
[███████████████████████████-------------] 66% [01m:06s] Processing chr. 22
[███████████████████████████-------------] 67% [01m:07s] Processing chr. 22
[████████████████████████████------------] 68% [01m:08s] Processing chr. 22
[████████████████████████████------------] 69% [01m:09s] Processing chr. 22
[█████████████████████████████-----------] 70% [01m:10s] Processing chr. 22
[█████████████████████████████-----------] 71% [01m:11s] Processing chr. 22
[█████████████████████████████-----------] 72% [01m:12s] Processing chr. 22
[██████████████████████████████----------] 73% [01m:13s] Processing chr. 22
[██████████████████████████████----------] 74% [01m:14s] Processing chr. 22
[███████████████████████████████---------] 75% [01m:15s] Processing chr. 22
[███████████████████████████████---------] 76% [01m:16s] Processing chr. 22
[███████████████████████████████---------] 77% [01m:17s] Processing chr. 22
[████████████████████████████████--------] 78% [01m:18s] Processing chr. 22
[████████████████████████████████--------] 79% [01m:19s] Processing chr. 22
[█████████████████████████████████-------] 80% [01m:20s] Processing chr. 22
[█████████████████████████████████-------] 81% [01m:21s] Processing chr. 22
[█████████████████████████████████-------] 82% [01m:22s] Processing chr. 22
[██████████████████████████████████------] 84% [01m:23s] Processing chr. 22
[███████████████████████████████████-----] 85% [01m:24s] Processing chr. 22
[███████████████████████████████████-----] 86% [01m:25s] Processing chr. 22
[███████████████████████████████████-----] 87% [01m:26s] Processing chr. 22
[████████████████████████████████████----] 88% [01m:27s] Processing chr. 22
[████████████████████████████████████----] 89% [01m:28s] Processing chr. 22
[█████████████████████████████████████---] 90% [01m:29s] Processing chr. 22
[█████████████████████████████████████---] 91% [01m:30s] Processing chr. 22
[█████████████████████████████████████---] 92% [01m:31s] Processing chr. 22
[██████████████████████████████████████--] 93% [01m:32s] Processing chr. 22
[██████████████████████████████████████--] 94% [01m:33s] Processing chr. 22
[███████████████████████████████████████-] 95% [01m:34s] Processing chr. 22
[███████████████████████████████████████-] 96% [01m:35s] Processing chr. 22
[███████████████████████████████████████-] 97% [01m:36s] Processing chr. 22
[████████████████████████████████████████] 98% [01m:37s] Processing chr. 22
[████████████████████████████████████████] 100% [01m:39s] Reads simulated
library(dplyr)
# filter germinal mutations
f_seq <- seq_results$mutations %>% dplyr::filter(classes!="germinal")
# plotting the VAF histogram
plot_VAF_histogram(f_seq, cuts = c(0.02, 1))
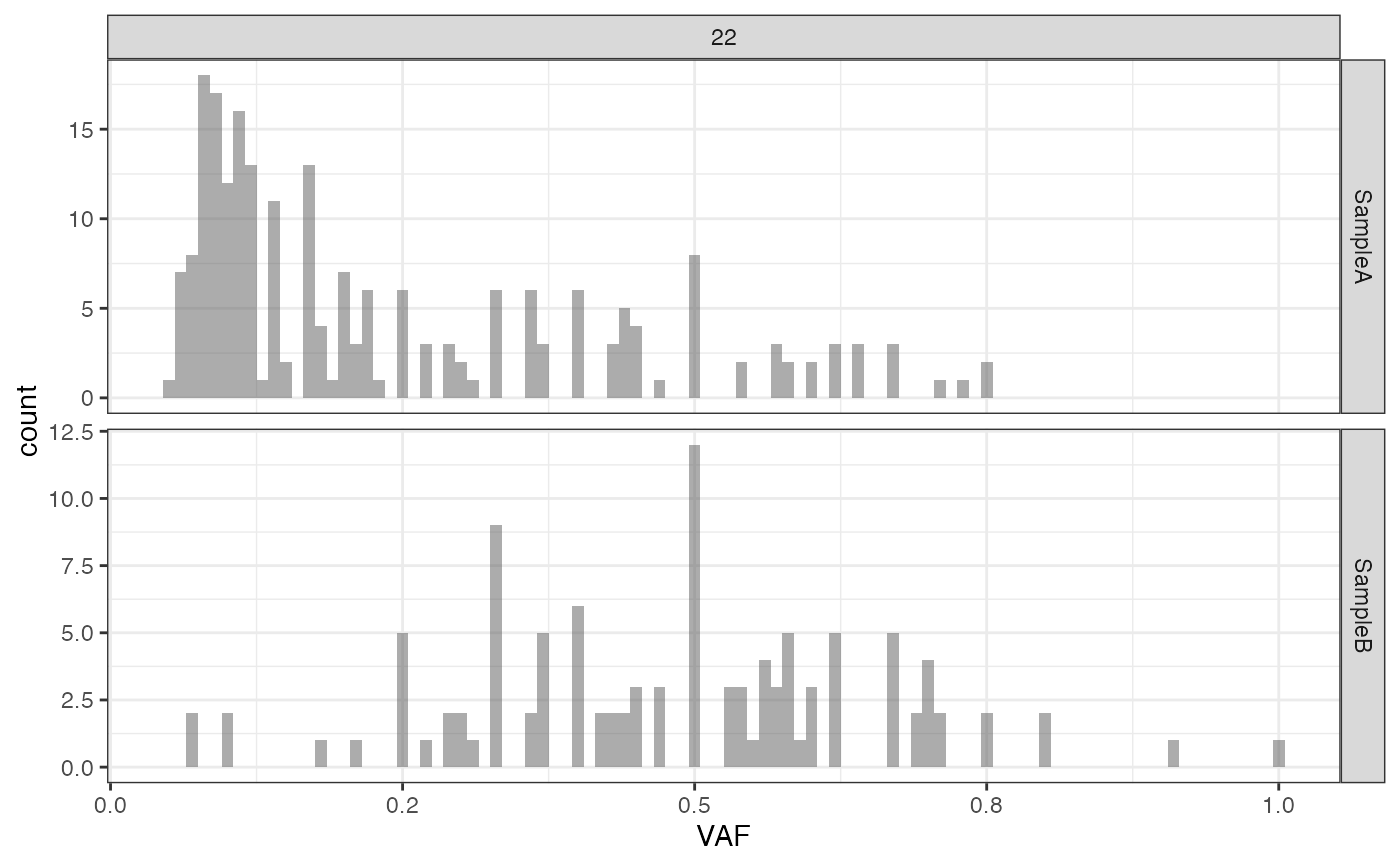 # plotting the VAF histogram with labels
plot_VAF_histogram(f_seq, labels = f_seq["causes"], cuts = c(0.02, 1))
# plotting the VAF histogram with labels
plot_VAF_histogram(f_seq, labels = f_seq["causes"], cuts = c(0.02, 1))
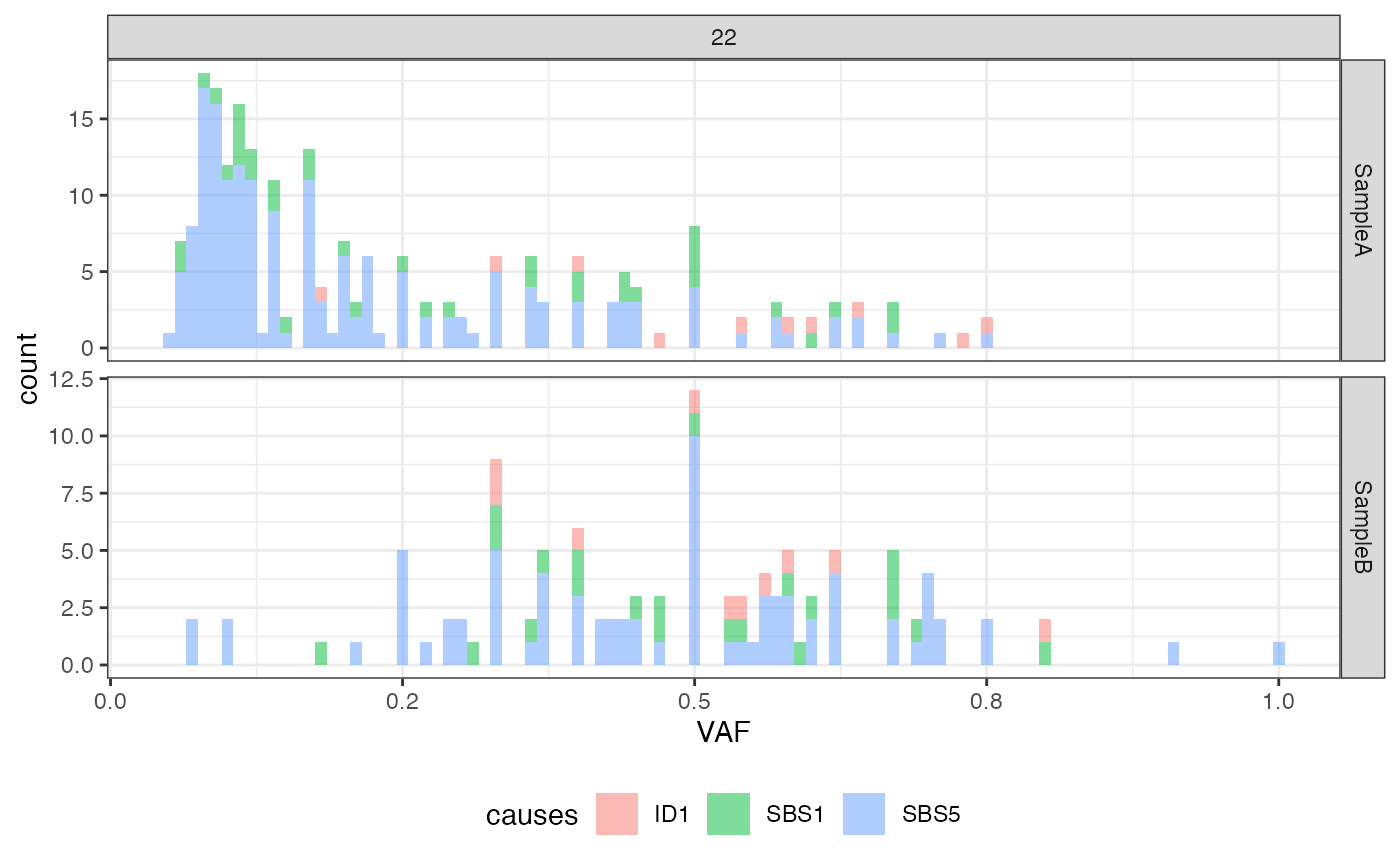 # deleting the mutation engine directory
unlink('demo', recursive = T)
# deleting the mutation engine directory
unlink('demo', recursive = T)