ctree
ctree.Rmd
# To render in colour this vignette
old.hooks <- fansi::set_knit_hooks(knitr::knit_hooks)The ctree is a package to implement basic functions to create, manipulate and visualize clone trees. A clone tree is a tree built from the results of a subclonal deconvolution analysis of bulk DNA sequencing data. The trees created with ctree are used inside REVOLVER, a package that implements one algorithm to determine repeated cancer evolution from multi-region sequencing data of human cancers.
To build a clone tree you need some basic datal; an example dataset is attached to the package and can be used to create a ctree S3 object.
data('ctree_input')Required data
Cancer Cell Fractions
Cancer Cell Fractions (CCF) clusters obtained from subclonal deconvolution analysis of bulk DNA sequencing data are required,
ctree_input$CCF_clusters#> # A tibble: 7 × 7
#> cluster nMuts is.driver is.clonal R1 R2 R3
#> <chr> <int> <lgl> <lgl> <dbl> <dbl> <dbl>
#> 1 1 72 TRUE FALSE 0 0.92 0
#> 2 2 69 TRUE TRUE 0.99 0.98 0.99
#> 3 3 48 FALSE FALSE 0 0 0.49
#> 4 4 29 FALSE FALSE 0.01 0.01 0.93
#> 5 5 24 TRUE FALSE 0.78 0 0
#> 6 6 23 TRUE FALSE 0.98 0.03 0.98
#> 7 7 15 FALSE FALSE 0 0.41 0If you need tools to compute CCF values, the evoverse collection of packages for Cancer Evolution analysis contains both MOBSTER and VIBER. Otherwise, a number of other packages can be used (pyClone, sciClone, DPClust, etc.).
Driver events mapped to the CCF clusters, with reported clonality status, a variantID and a patientID.
ctree_input$drivers#> # A tibble: 7 × 8
#> patientID variantID is.driver is.clonal cluster R1 R2 R3
#> <chr> <chr> <lgl> <lgl> <chr> <dbl> <dbl> <dbl>
#> 1 CRUK0002 RB1 TRUE FALSE 1 0 0.92 0
#> 2 CRUK0002 IKZF1 TRUE FALSE 1 0 0.92 0
#> 3 CRUK0002 KRAS TRUE FALSE 1 0 0.93 0
#> 4 CRUK0002 MET TRUE TRUE 2 0.99 0.98 0.99
#> 5 CRUK0002 TERT TRUE TRUE 2 0.99 0.98 0.99
#> 6 CRUK0002 NF1 TRUE FALSE 5 0.78 0 0
#> 7 CRUK0002 EP300 TRUE FALSE 6 0.96 0.03 0.98Other data
ctree_input$samples
#> [1] "R1" "R2" "R3"
ctree_input$patient
#> [1] "CUK12345"Creation of a clone tree
You can use a sampler and control its parameters – see ~ctrees.
x = ctrees(
CCF_clusters = ctree_input$CCF_clusters,
drivers = ctree_input$drivers,
samples = ctree_input$samples,
patient = ctree_input$patient,
sspace.cutoff = ctree_input$sspace.cutoff,
n.sampling = ctree_input$n.sampling,
store.max = ctree_input$store.max
)#> [ ctree ~ clone trees generator for CUK12345 ]
#>
#> # A tibble: 7 × 7
#> cluster nMuts is.driver is.clonal R1 R2 R3
#> <chr> <int> <lgl> <lgl> <dbl> <dbl> <dbl>
#> 1 1 72 TRUE FALSE 0 0.92 0
#> 2 2 69 TRUE TRUE 0.99 0.98 0.99
#> 3 3 48 FALSE FALSE 0 0 0.49
#> 4 4 29 FALSE FALSE 0.01 0.01 0.93
#> 5 5 24 TRUE FALSE 0.78 0 0
#> 6 6 23 TRUE FALSE 0.98 0.03 0.98
#> 7 7 15 FALSE FALSE 0 0.41 0#> ✔ Trees per region 1, 3, 1
#> ℹ Total 3 tree structures - search is exahustive
#>
#> ── Ranking trees
#> ✔ 3 trees with non-zero score, storing 3The sampler creates a number of clone trees that can fit the data according to an error model that allows violations of the pigeonhole principle. We work with the top-ranking model.
x = x[[1]]Visualisations
S3 functions for printing, and summarizing the object.
print(x)#> [ ctree - ctree rank 1/3 for CUK12345 ]
#>
#> # A tibble: 7 × 7
#> cluster nMuts is.driver is.clonal R1 R2 R3
#> <chr> <int> <lgl> <lgl> <dbl> <dbl> <dbl>
#> 1 1 72 TRUE FALSE 0 0.92 0
#> 2 2 69 TRUE TRUE 0.99 0.98 0.99
#> 3 3 48 FALSE FALSE 0 0 0.49
#> 4 4 29 FALSE FALSE 0.01 0.01 0.93
#> 5 5 24 TRUE FALSE 0.78 0 0
#> 6 6 23 TRUE FALSE 0.98 0.03 0.98
#> 7 7 15 FALSE FALSE 0 0.41 0
#>
#> Tree shape (drivers annotated)
#>
#> \-GL
#> \-2 :: MET, TERT
#> |-1 :: RB1, IKZF1, KRAS
#> | \-7
#> \-6 :: EP300
#> |-4
#> | \-3
#> \-5 :: NF1
#>
#> Information transfer
#>
#> MET ---> RB1
#> MET ---> IKZF1
#> MET ---> KRAS
#> TERT ---> RB1
#> TERT ---> IKZF1
#> TERT ---> KRAS
#> GL ---> MET
#> GL ---> TERT
#> EP300 ---> NF1
#> MET ---> EP300
#> TERT ---> EP300
#>
#> Tree score 0.6
#>
summary(x)#> [ ctree - ctree rank 1/3 for CUK12345 ]
#>
#> # A tibble: 7 × 7
#> cluster nMuts is.driver is.clonal R1 R2 R3
#> <chr> <int> <lgl> <lgl> <dbl> <dbl> <dbl>
#> 1 1 72 TRUE FALSE 0 0.92 0
#> 2 2 69 TRUE TRUE 0.99 0.98 0.99
#> 3 3 48 FALSE FALSE 0 0 0.49
#> 4 4 29 FALSE FALSE 0.01 0.01 0.93
#> 5 5 24 TRUE FALSE 0.78 0 0
#> 6 6 23 TRUE FALSE 0.98 0.03 0.98
#> 7 7 15 FALSE FALSE 0 0.41 0
#>
#> Tree shape (drivers annotated)
#>
#> \-GL
#> \-2 :: MET, TERT
#> |-1 :: RB1, IKZF1, KRAS
#> | \-7
#> \-6 :: EP300
#> |-4
#> | \-3
#> \-5 :: NF1
#>
#> Information transfer
#>
#> MET ---> RB1
#> MET ---> IKZF1
#> MET ---> KRAS
#> TERT ---> RB1
#> TERT ---> IKZF1
#> TERT ---> KRAS
#> GL ---> MET
#> GL ---> TERT
#> EP300 ---> NF1
#> MET ---> EP300
#> TERT ---> EP300
#>
#> Tree score 0.6
#>
#> CCF clusters:
#>
#> # A tibble: 7 × 7
#> cluster nMuts is.driver is.clonal R1 R2 R3
#> <chr> <int> <lgl> <lgl> <dbl> <dbl> <dbl>
#> 1 1 72 TRUE FALSE 0 0.92 0
#> 2 2 69 TRUE TRUE 0.99 0.98 0.99
#> 3 3 48 FALSE FALSE 0 0 0.49
#> 4 4 29 FALSE FALSE 0.01 0.01 0.93
#> 5 5 24 TRUE FALSE 0.78 0 0
#> 6 6 23 TRUE FALSE 0.98 0.03 0.98
#> 7 7 15 FALSE FALSE 0 0.41 0
#>
#> Drivers:
#>
#> # A tibble: 7 × 8
#> patientID variantID is.driver is.clonal cluster R1 R2 R3
#> <chr> <chr> <lgl> <lgl> <chr> <dbl> <dbl> <dbl>
#> 1 CRUK0002 RB1 TRUE FALSE 1 0 0.92 0
#> 2 CRUK0002 IKZF1 TRUE FALSE 1 0 0.92 0
#> 3 CRUK0002 KRAS TRUE FALSE 1 0 0.93 0
#> 4 CRUK0002 MET TRUE TRUE 2 0.99 0.98 0.99
#> 5 CRUK0002 TERT TRUE TRUE 2 0.99 0.98 0.99
#> 6 CRUK0002 NF1 TRUE FALSE 5 0.78 0 0
#> 7 CRUK0002 EP300 TRUE FALSE 6 0.96 0.03 0.98
#>
#> Pigeonhole principle: 12 0
#>
#> R1 R2 R3
#> 1 TRUE TRUE TRUE
#> 2 TRUE TRUE TRUE
#> 4 TRUE TRUE TRUE
#> 6 TRUE TRUE TRUE
#>
#> Goodness-of-fit: 1
#>
#> Then some plotting functions for the tree. A tree layout is used to display the clone tree and the information transfer, which corresponds to the ordering of the drivers annnotated in the tree. This terminology is borrowed from the REVOLVER algorithm, where it is used to refer to the set of trajectories that a patient “transfers” to another patient during the fit.
plot(x)
#> Warning: Duplicated aesthetics after name standardisation: na.rm
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> =
#> "none")` instead.
#> Warning: Removed 1 rows containing missing values (geom_point).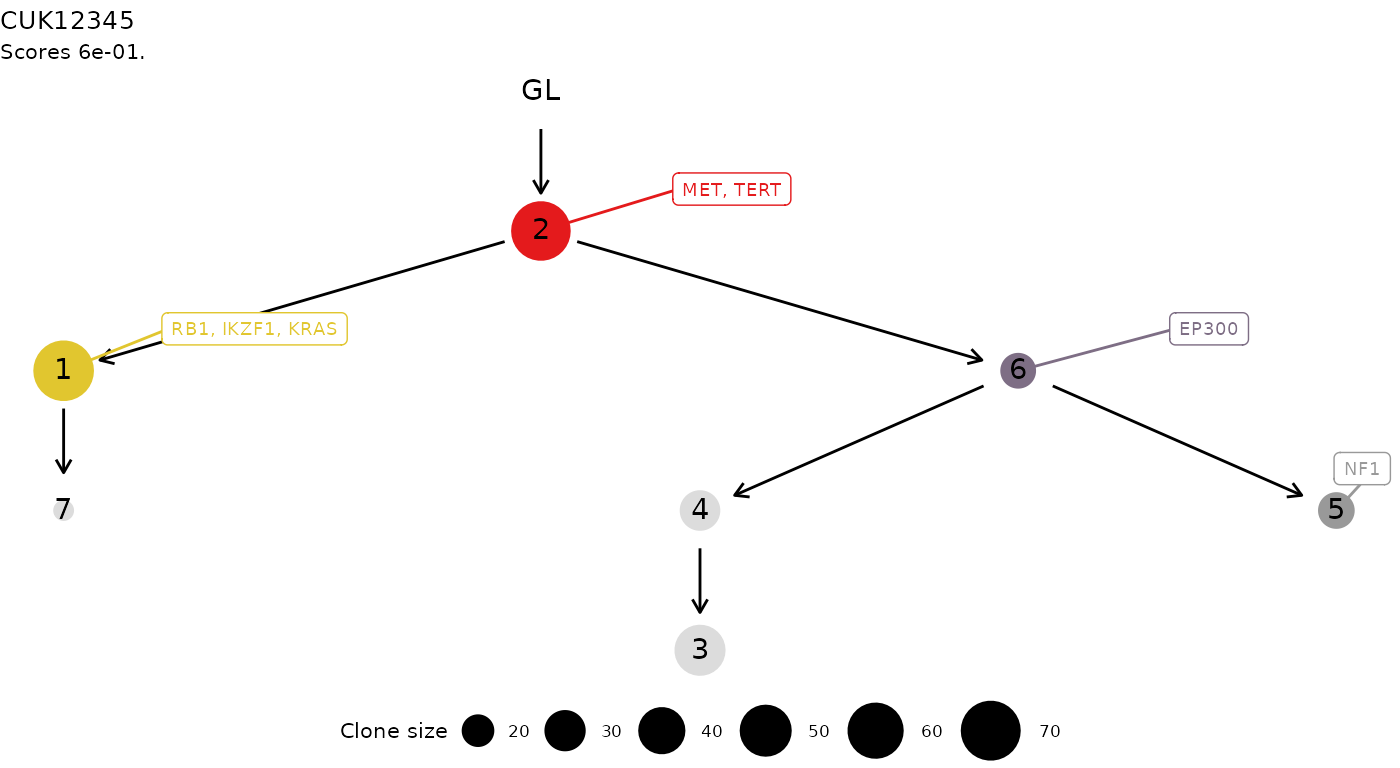
plot_information_transfer(x)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> =
#> "none")` instead.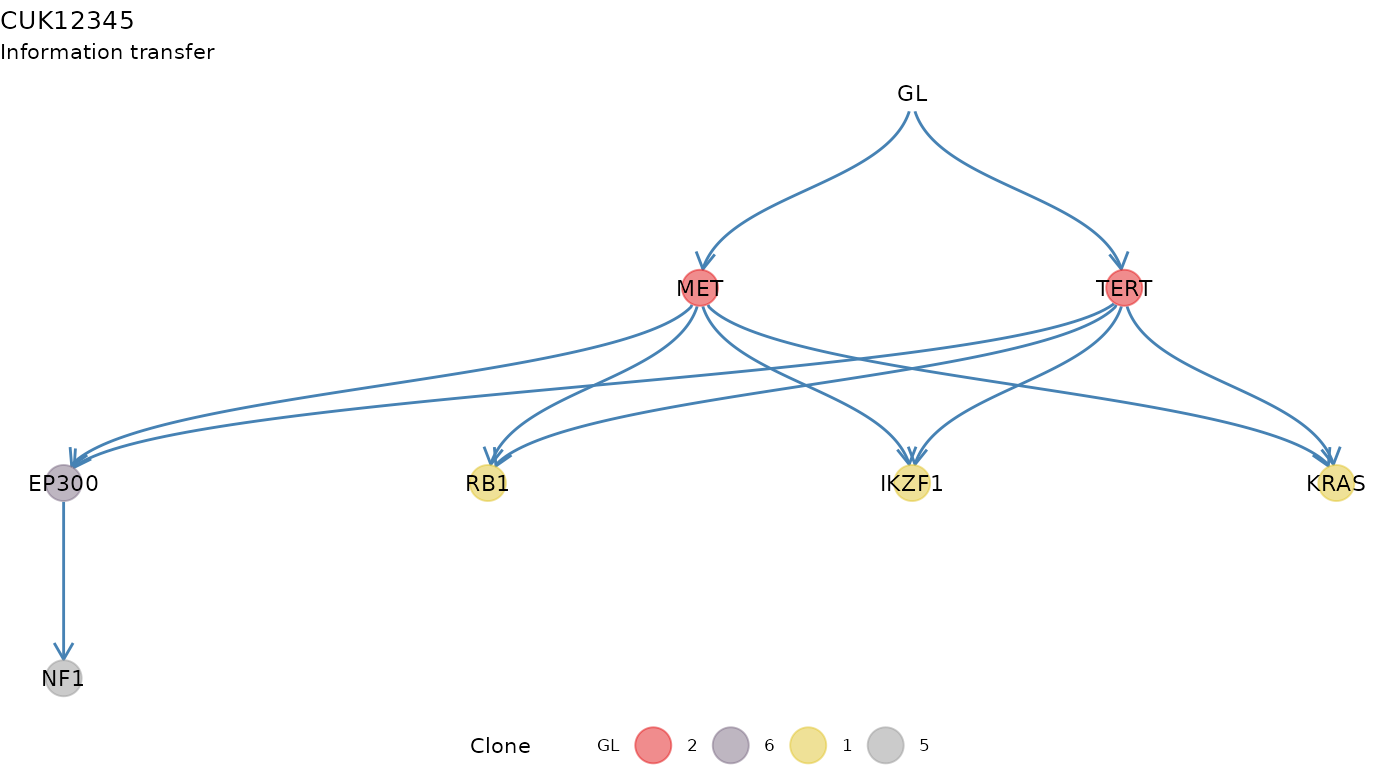
plot_icon(x)
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> =
#> "none")` instead.
For the data, you can plot the CCF of the clusters.
plot_CCF_clusters(x)
#> Warning: Removed 8 rows containing missing values (geom_text).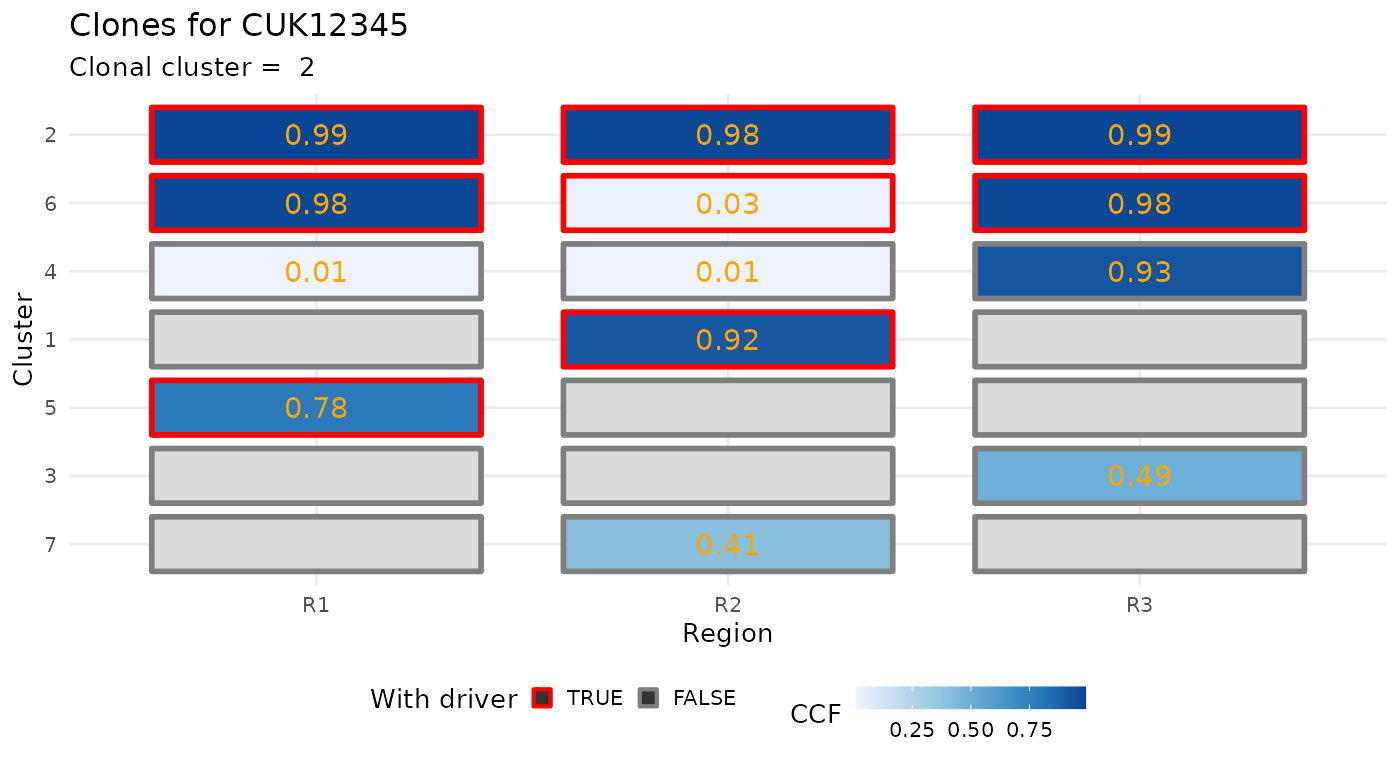
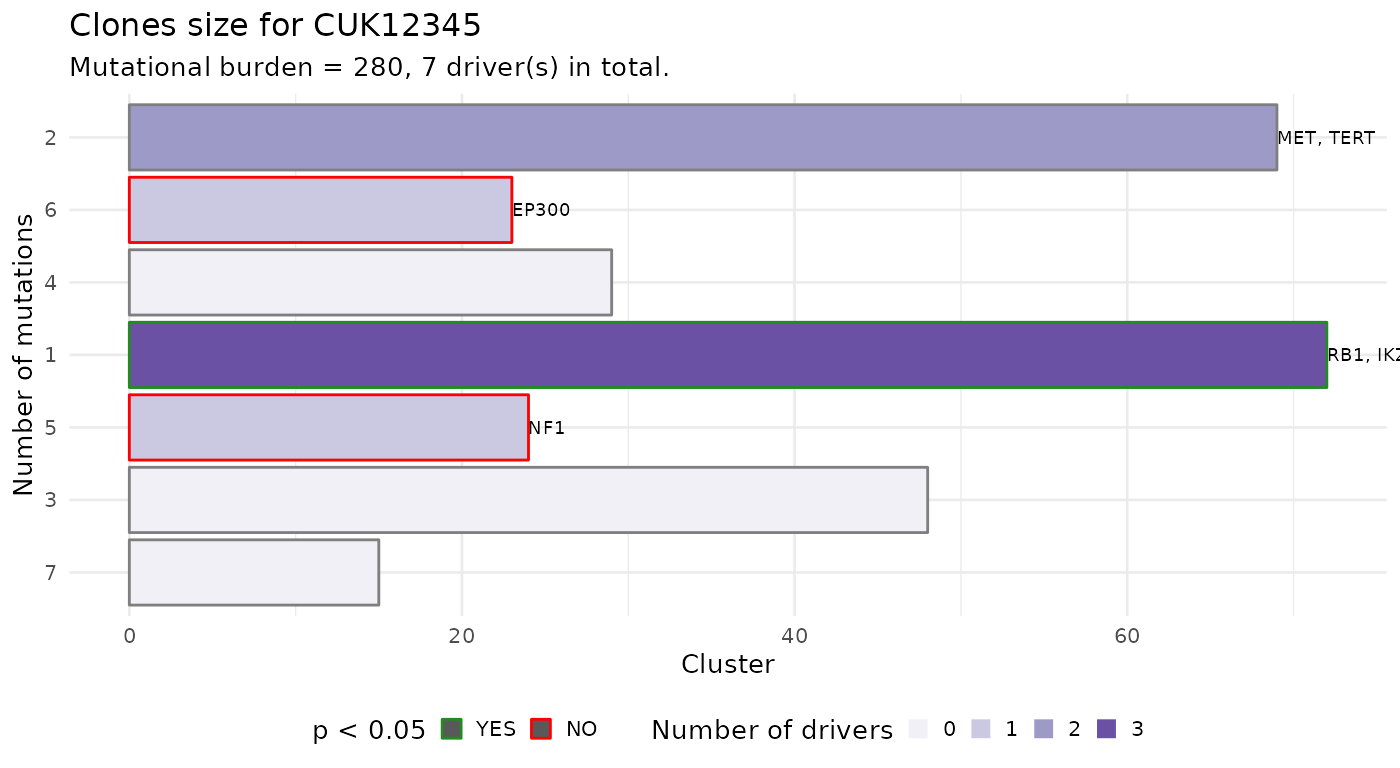
Or you can plot the size of each CCF cluster as a barplot. This barplot is annotated to report wether a subclone with a driver is significantly larger than the expected size for a subclone without driver. To carry out this test subclones without drivers are used to estimate the parameters of a univariate Gaussian distribution (mean and standard deviation), the p-value is then computed from the fit distribution through the pnorm function. The confidence level for the test can be passed as parameter.
plot_clone_size(x)
#> Warning: Removed 3 rows containing missing values (geom_text).