4. Fitting a mixture of resistant-sensitive population
a4_task3.Rmd
library(biPOD)
require(dplyr)
#> Loading required package: dplyr
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, unionWhen a population shrinks (e.g., sensitive to treatment) and later regrows (e.g., resistant to treatment) the dynamics are U-shaped. Here we are going to consider this specific case.
Input data
We take the xenografts data and we make it ready for
inference by preparing the count column and by dividing the
time column by a factor of 7 (in order to work with a unit
of time of a week).
We select the sample 543, already presented in the previous vignette and we filter out the observations that are prior to the treatment.
data("xenografts", package = "biPOD")
mouse_id <- 543
d <- xenografts %>%
dplyr::rename(count = tumour_volume) %>%
dplyr::mutate(time = time / 7) %>%
dplyr::filter(time >= 0) %>%
dplyr::filter(mouse == mouse_id)
x <- biPOD::init(d, "543 U-shape")
#>
#> ── biPOD - bayesian inference for Population Dynamics ──────────────────────────
#> ℹ Using sample named: 543 U-shape.
#> ! No group column present in input dataframe! A column will be added.
biPOD::plot_input(x)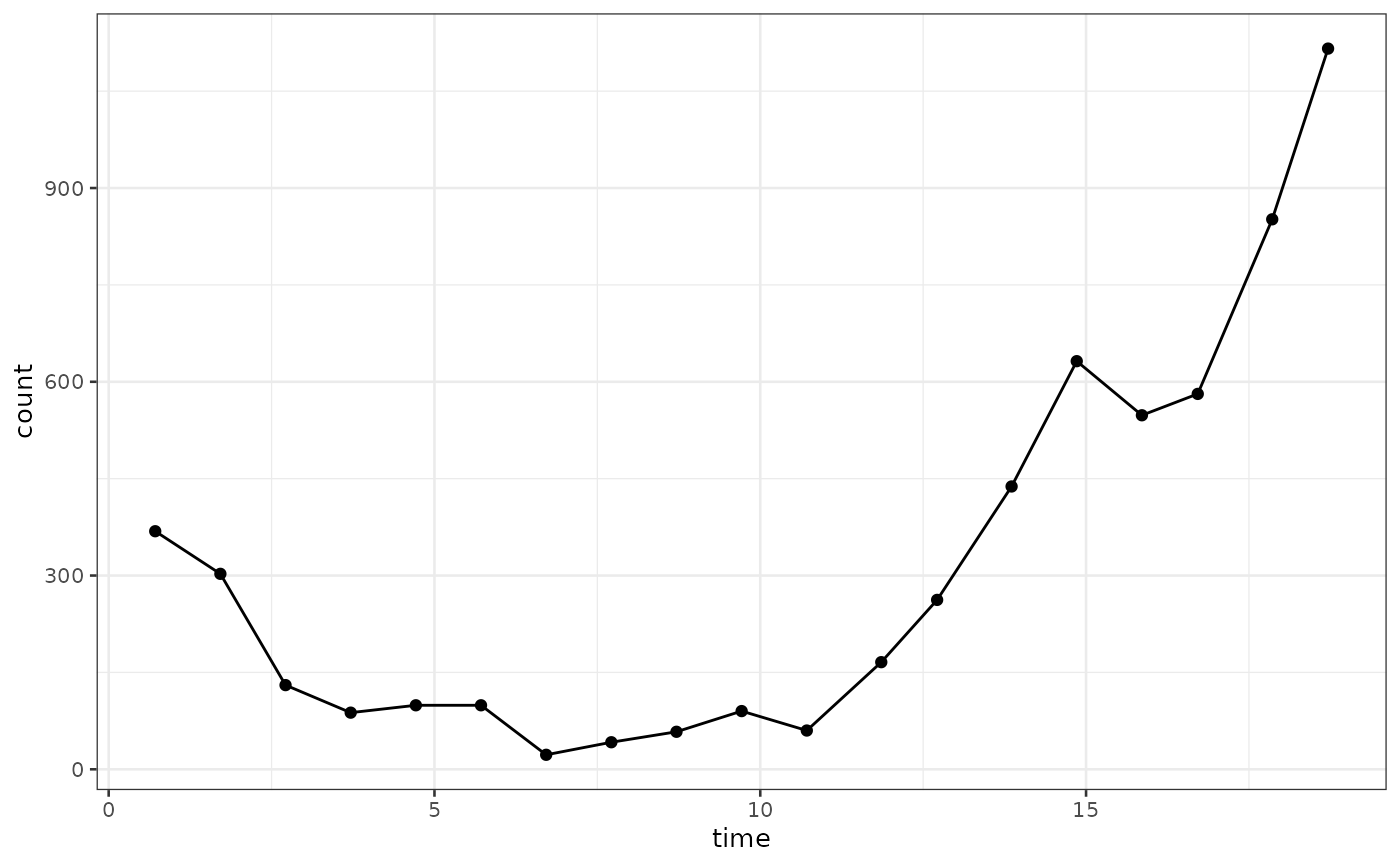
Two population model
The model assumes that
- only two populations exist, one dying out and one growing
- the growth rates are both positive and constant
- the sensible population must be present at x=0 but the same does not apply for the resistant one
and is able to infer
- , instant of time in which the sensitive population dies out
- , instant of time in which the resistant population was born
- , death rate of the sensitive population
- , growth rate of the resistant population
To do so, one need to use the fit_two_pop_model
function.
x <- biPOD::fit_two_pop_model(x, variational = F, factor_size = 1)
#> ℹ Fitting two population model using MCMC sampling ...
#> Chain 4 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 4 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe5298e1028.stan', line 29, column 2 to column 18)
#> Chain 4 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 4 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 4
#> Chain 1 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 1 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 1 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 1 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 1
#> Chain 1 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 1 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 1 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 1 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 1
#> Chain 1 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 1 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 1 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 1 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 1
#> Chain 3 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 3 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 3 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 3 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 3
#> Chain 3 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 3 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 3 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 3 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 3
#> Chain 3 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 3 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 3 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 3 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 3
#> Chain 3 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 3 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 3 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 3 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 3
#> Chain 3 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:
#> Chain 3 Exception: poisson_lpmf: Rate parameter[1] is -nan, but must be nonnegative! (in '/tmp/RtmpmcIZny/model-1fe51dd37129.stan', line 44, column 2 to column 18)
#> Chain 3 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,
#> Chain 3 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.
#> Chain 3
#> Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.
#> Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.
#> Warning: Some Pareto k diagnostic values are too high. See help('pareto-k-diagnostic') for details.
#> Warning: Dropping 'draws_df' class as required metadata was removed.
#> Warning: Dropping 'draws_df' class as required metadata was removed.
#> Warning: Dropping 'draws_df' class as required metadata was removed.
#> Warning: Dropping 'draws_df' class as required metadata was removed.
#> Warning: Dropping 'draws_df' class as required metadata was removed.
#> Warning: Dropping 'draws_df' class as required metadata was removed.
#> Warning: Dropping 'draws_df' class as required metadata was removed.and the fit can be visualized as a single process
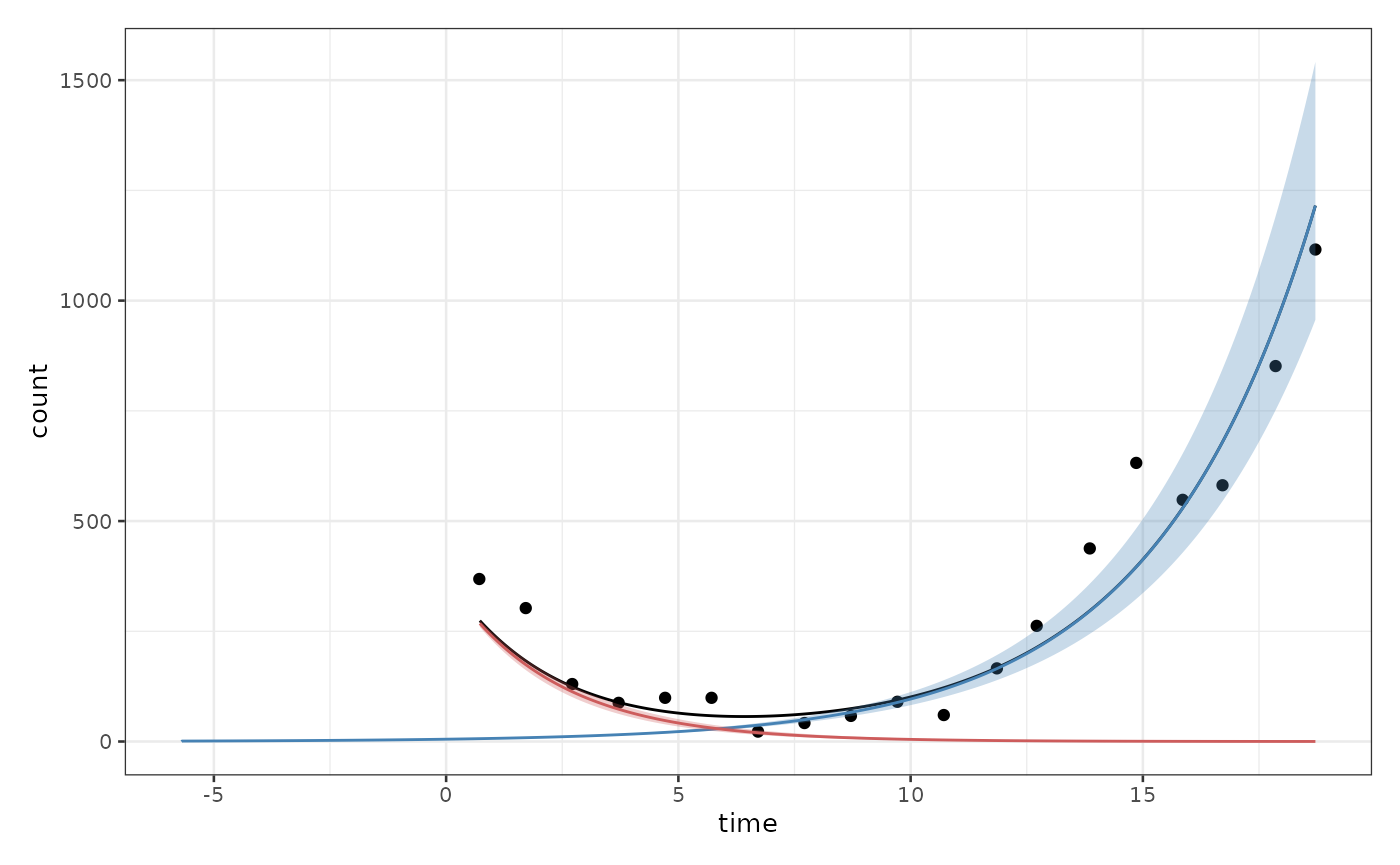
biPOD::plot_two_pop_fit(x, split_process = F, f_posteriors = F, t_posteriors = F, r_posteriors = F)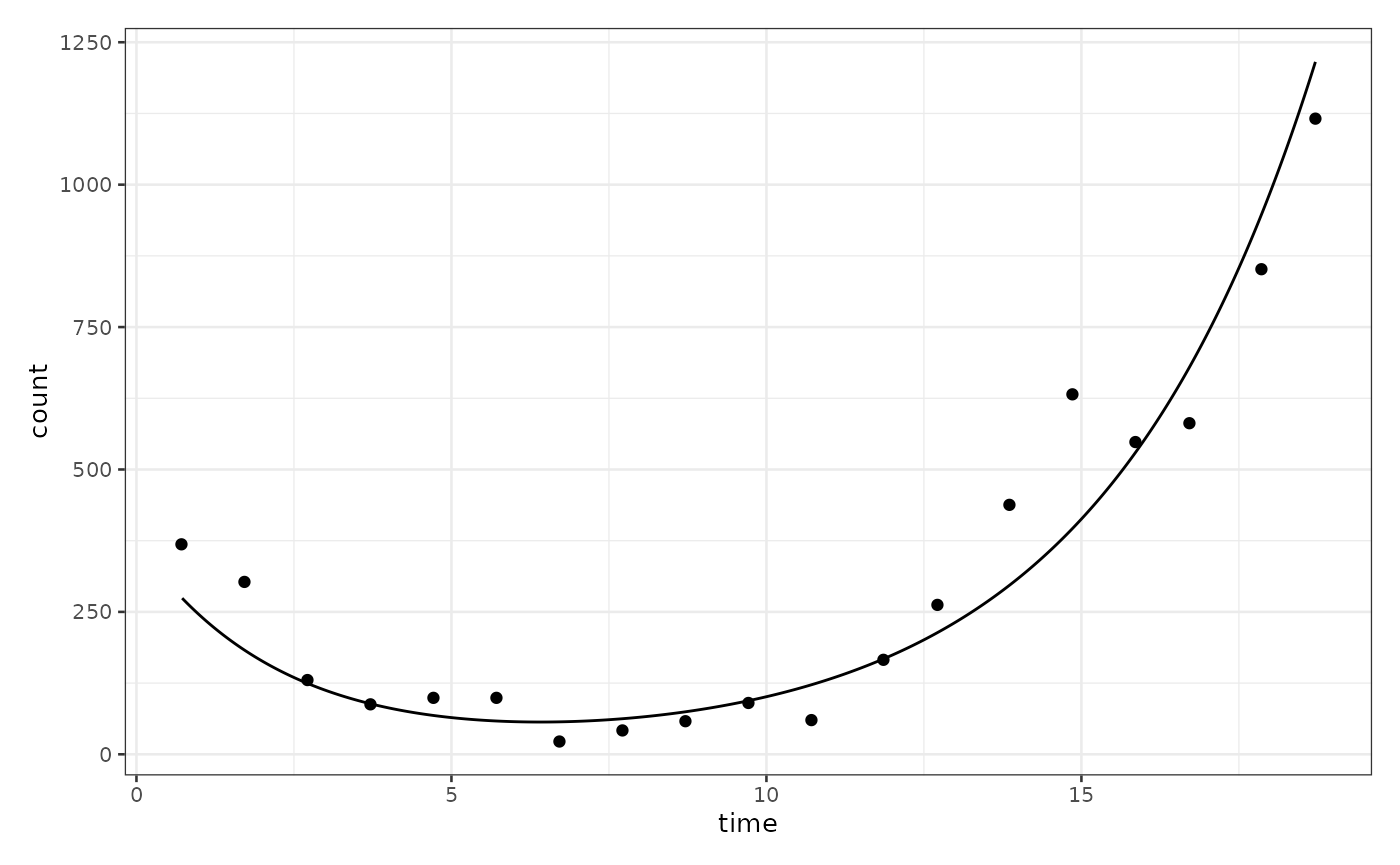
or splitting it depending on the two different populations
biPOD::plot_two_pop_fit(x, split_process = T, f_posteriors = F, t_posteriors = F, r_posteriors = F)