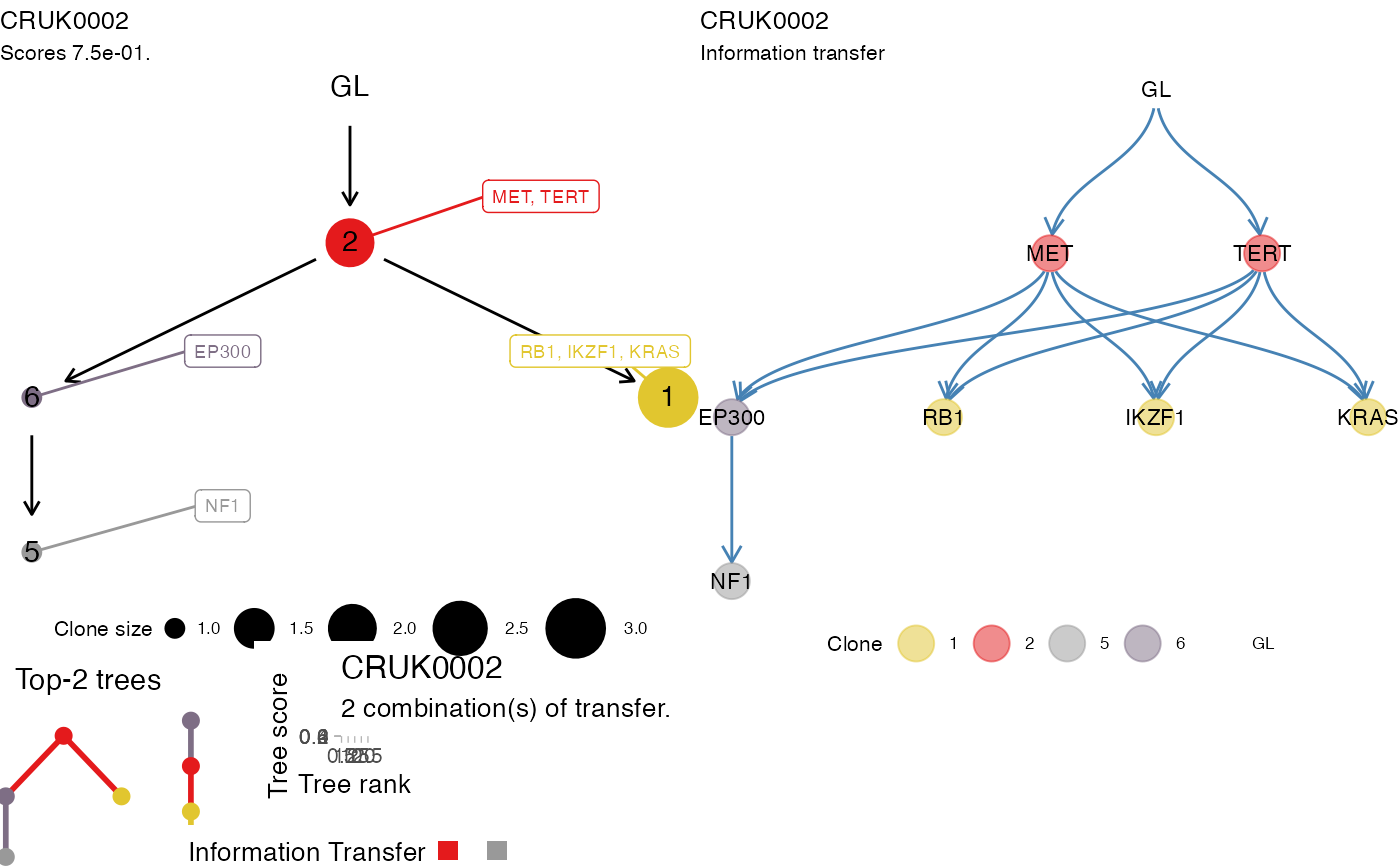
This function is like plot_data, as it uses base plotting
functions to assemble a summary plot for a patient. This function
assembled the plot via ggpubr. The first line of plots
represents the top tree for a patient, and its information transfer.
The strip below represents up to the top-10 trees for this patient,
as they are obtained from the standard tree-scoring (which means
that the score is not accounting for the actual transfer, but just
for the tree structure).
plot_patient_trees(x, patient, ...)
Arguments
| x | A REVOLVER cohort object |
|---|---|
| patient | The patient for which the trees should be plot |
| ... | Extra parameters, not used. |
Value
A figure assembled with ggpubr.
Examples
# Data released in the 'evoverse.datasets' data('TRACERx_NEJM_2017_REVOLVER', package = 'evoverse.datasets') # This returns a figure assembled with ggpubr plot_patient_trees(TRACERx_NEJM_2017_REVOLVER, patient = 'CRUK0002')#> Warning: Duplicated aesthetics after name standardisation: na.rm#> Warning: Removed 1 rows containing missing values (geom_point).