This function uses the output fit of VIBER
to create a call to ctree (https://caravagn.github.io/ctree/),
a package to create clone trees for cancer evolution models.
Creation of a clone tree requires annotations that are not usually
necessary for just a plain VIBER analyses. These annotations report the status of driver
and gene for each one of the input datapoints; the annotations should
have been passed when calling the variational_fit function, and stored
inside the data field of the VIBER object.
The clonal cluster is estimated from the cluster with the highest parameter values in most of the input dimensions (Binomial peaks).
The output is the result of calling the constructor ctree::cetrees
on the input clustering results x.
get_clone_trees(x, ...)Arguments
- x
A VIBER fit.
- ...
Extra parameters passed to the constructor
ctree::cetrees, which affect the sampling of the trees.
Value
The output of the constructor ctree::cetrees.
Examples
data(mvbmm_example)
# We create annotation data assigning dummy names
# and picking 10 events to be drivers (randomly chosen)
data_annotations = data.frame(
gene = paste0("G", 1:nrow(mvbmm_example$trials)),
driver = FALSE
)
data_annotations$driver[sample(1:nrow(data_annotations), 10)] = TRUE
# Compared to the main variational_fit, we use the same call but add data
f = variational_fit(mvbmm_example$successes, mvbmm_example$trials, data = data_annotations)
#> [ VIBER - variational fit ]
#>
#> ℹ Input n = 231, with k < 10. Dirichlet concentration α = 1e-06.
#> ℹ Beta (a_0, b_0) = (1, 1); q_i = prior. Optimise: ε = 1e-10 or 5000 steps, r = 10 starts.
#>
#> ✔ VIBER fit completed in 0.1 mins (status: converged)
#>
#> ── [ VIBER ] My VIBER model n = 231 (w = 2 dimensions). Fit with k = 10 clusters
#> • Clusters: π = 45% [C9], 28% [C2], 20% [C4], and 7% [C5], with π > 0.
#> • Binomials: θ = <0.5, 0.49> [C9], <0, 0.2> [C2], <0.25, 0.25> [C4], and <0.22,
#> 0> [C5].
#> ℹ Score(s): ELBO = -47073.317. Fit converged in 24 steps, ε = 1e-10.
print(f)
#> ── [ VIBER ] My VIBER model n = 231 (w = 2 dimensions). Fit with k = 10 clusters
#> • Clusters: π = 45% [C9], 28% [C2], 20% [C4], and 7% [C5], with π > 0.
#> • Binomials: θ = <0.5, 0.49> [C9], <0, 0.2> [C2], <0.25, 0.25> [C4], and <0.22,
#> 0> [C5].
#> ℹ Score(s): ELBO = -47073.317. Fit converged in 24 steps, ε = 1e-10.
trees = get_clone_trees(f)
#> Estimated clonal cluster C9 from VIBER fit.
#> Found 3 driver event(s) in VIBER fits.
#> [ ctree ~ clone trees generator for VIBER_dataset ]
#>
#> # A tibble: 4 × 6
#> cluster S1 S2 nMuts is.clonal is.driver
#> <chr> <dbl> <dbl> <int> <lgl> <lgl>
#> 1 C2 0.000155 0.203 65 FALSE TRUE
#> 2 C4 0.251 0.254 47 FALSE FALSE
#> 3 C5 0.217 0.000657 16 FALSE TRUE
#> 4 C9 0.498 0.493 103 TRUE TRUE
#>
#> ✔ Trees per region 2, 2
#> ℹ Total 4 tree structures - search is exahustive
#>
#> ── Ranking trees
#> ✔ 4 trees with non-zero score, storing 4
ctree:::print.ctree(trees[[1]])
#> [ ctree - ctree rank 1/4 for VIBER_dataset ]
#>
#> # A tibble: 4 × 6
#> cluster S1 S2 nMuts is.clonal is.driver
#> <chr> <dbl> <dbl> <int> <lgl> <lgl>
#> 1 C2 0.000155 0.203 65 FALSE TRUE
#> 2 C4 0.251 0.254 47 FALSE FALSE
#> 3 C5 0.217 0.000657 16 FALSE TRUE
#> 4 C9 0.498 0.493 103 TRUE TRUE
#>
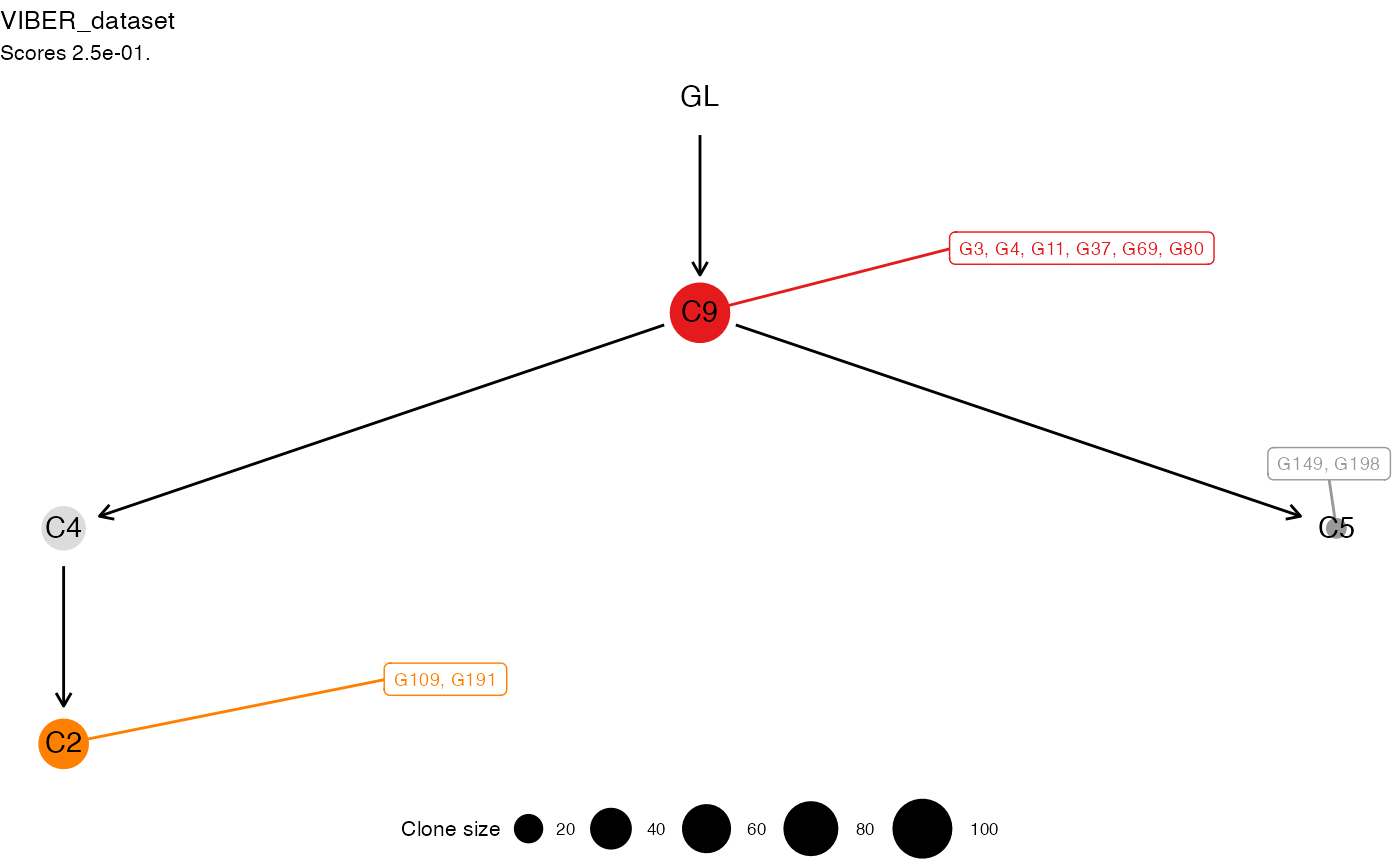
#> Tree shape (drivers annotated)
#>
#> \-GL
#> \-C9 :: G3, G4, G11, G37, G69, G80
#> |-C4
#> | \-C2 :: G109, G191
#> \-C5 :: G149, G198
#>
#> Information transfer
#>
#> G3 ---> G109
#> G3 ---> G191
#> G4 ---> G109
#> G4 ---> G191
#> G11 ---> G109
#> G11 ---> G191
#> G37 ---> G109
#> G37 ---> G191
#> G69 ---> G109
#> G69 ---> G191
#> G80 ---> G109
#> G80 ---> G191
#> G3 ---> G149
#> G3 ---> G198
#> G4 ---> G149
#> G4 ---> G198
#> G11 ---> G149
#> G11 ---> G198
#> G37 ---> G149
#> G37 ---> G198
#> G69 ---> G149
#> G69 ---> G198
#> G80 ---> G149
#> G80 ---> G198
#> GL ---> G3
#> GL ---> G4
#> GL ---> G11
#> GL ---> G37
#> GL ---> G69
#> GL ---> G80
#>
#> Tree score 0.25
#>
ctree::plot.ctree(trees[[1]])
#> Warning: Duplicated aesthetics after name standardisation: na.rm
#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.
#> Warning: Removed 1 rows containing missing values (geom_point).