This function simulates a random dataset for TINC analysis,
with mutations and copy number data. Segments are not real, they are assumed
to be constant heterozygous diploid (Major = minor = 1) and span just
mutations (for mappability).
This samples has some noise so that the obtained TIT score might be slightly lower than the required input.
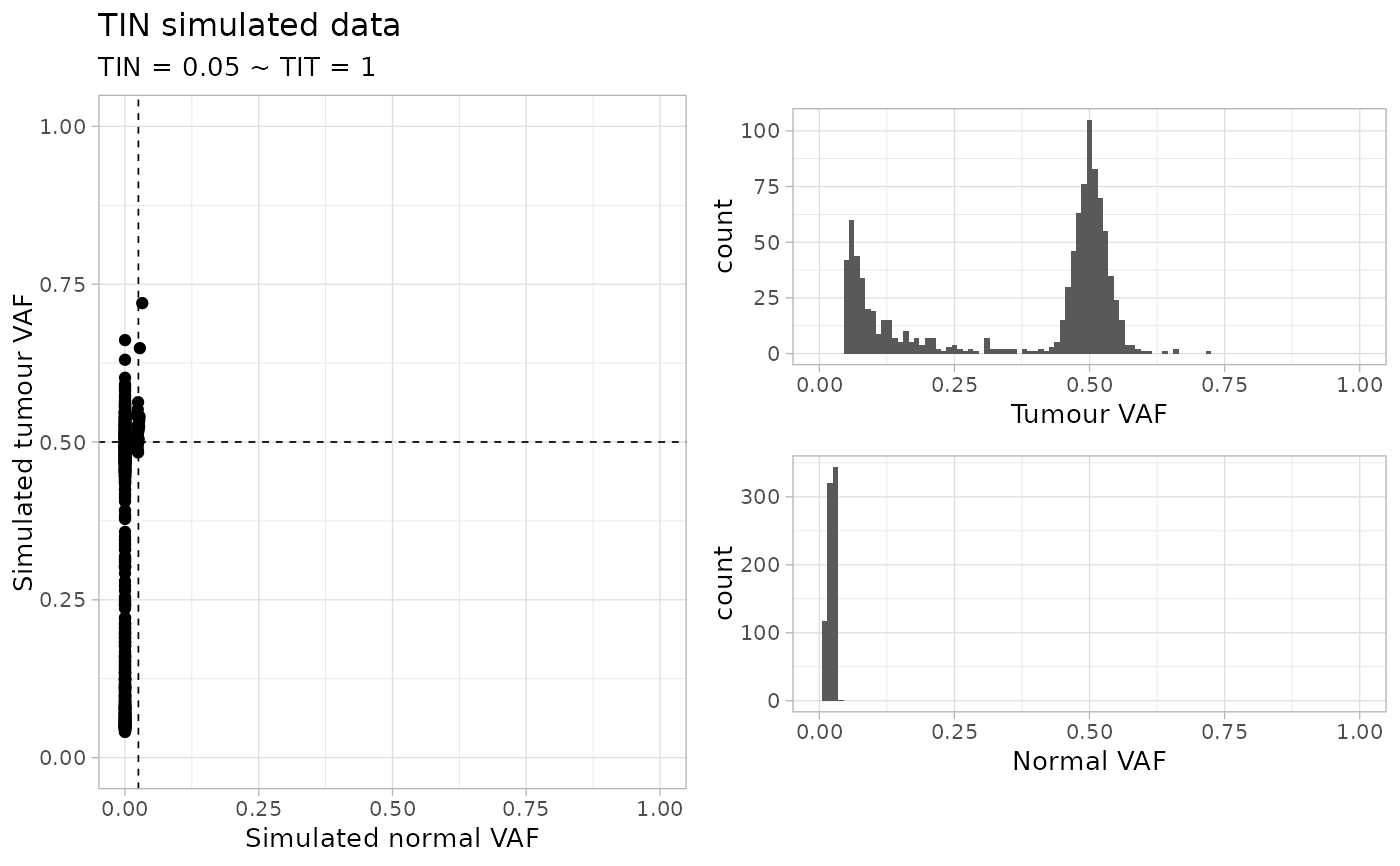
random_TIN(
N = 1000,
TIN = 0.05,
TIT = 1,
normal_coverage = 30,
tumour_coverage = 120
)Arguments
- N
Number of input simulations
- TIN
TIN - Tumour in normal contamination level.
- TIT
TIT - Tumour in tumour contamination level (aka tumour purity).
- normal_coverage
Normal coverage (mean).
- tumour_coverage
Tumour coverage (mean).
Value
Tibbles with the data, and a plot.
Examples
set.seed(1234)
# Default dataset
random_TIN()
#> ✔ Generated TINC dataset (n = 991 mutations), TIN (0.05) and TIT (1), normal and tumour coverage 30x and 120x.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
#> $data
#> # A tibble: 991 × 14
#> chr from to ref alt simulated_cluster n_tot_count t_tot_count
#> <chr> <int> <dbl> <chr> <chr> <chr> <int> <int>
#> 1 chr21 9350303 9.35e6 T C C1 35 109
#> 2 chr22 34134600 3.41e7 C A C1 40 127
#> 3 chr10 28158356 2.82e7 T C C1 37 120
#> 4 chr13 76030019 7.60e7 G A C1 43 113
#> 5 chr7 80111695 8.01e7 T C C1 27 107
#> 6 chr16 79722541 7.97e7 T T C1 42 122
#> 7 chr10 106104245 1.06e8 A T C1 28 122
#> 8 chr15 95490382 9.55e7 G G C1 24 124
#> 9 chr6 17255265 1.73e7 G A C1 32 120
#> 10 chr7 143422347 1.43e8 C C C1 43 135
#> # ℹ 981 more rows
#> # ℹ 6 more variables: n_alt_count <dbl>, t_alt_count <dbl>, n_ref_count <dbl>,
#> # t_ref_count <dbl>, sim_t_vaf <dbl>, sim_n_vaf <dbl>
#>
#> $cna
#> # A tibble: 991 × 6
#> chr from to ref Major minor
#> <chr> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 chr21 9350302 9350305 T 1 1
#> 2 chr22 34134599 34134602 C 1 1
#> 3 chr10 28158355 28158358 T 1 1
#> 4 chr13 76030018 76030021 G 1 1
#> 5 chr7 80111694 80111697 T 1 1
#> 6 chr16 79722540 79722543 T 1 1
#> 7 chr10 106104244 106104247 A 1 1
#> 8 chr15 95490381 95490384 G 1 1
#> 9 chr6 17255264 17255267 G 1 1
#> 10 chr7 143422346 143422349 C 1 1
#> # ℹ 981 more rows
#>
#> $plot
 #>
#>