
1. Prior distribution of mutation copy number and multiplicity from PCAWG
Source:vignettes/a1_priors_k_m.Rmd
a1_priors_k_m.Rmd
library(INCOMMON)
#> Warning: replacing previous import 'cli::num_ansi_colors' by
#> 'crayon::num_ansi_colors' when loading 'INCOMMON'
library(dplyr)
library(DT)The inference of copy number and multiplicity of a mutation from read counts only can be much of a hard task, especially in cases where the sample purity or the sequencing depth at the mutation site are low.
For this reason, INCOMMON allows using a prior distribution to improve classifications.
1.1 Empirical priors from PCAWG
For the inference of mutation copy number and multiplicity on a specific gene and in samples of a specific tumour type, a Dirichlet prior distribution , where is the total copy number and the mutation multiplicity, can be used to obtain more confident predictions, given that is obtained from reliable copy number calls. By default, INCOMMON relies on prior probability obtained from PCAWG and HMF whole genomes. From a set of high-confidence copy number calls validated by quality control, we obtained for each gene as the frequency of the corresponding INCOMMON class.
data("priors_pcawg_hmf")The empirical priors from PCAWG and HMF are provided as an internal
data table priors_pcawg_hmf and have the following
format
#> Warning in instance$preRenderHook(instance): It seems your data is too big for
#> client-side DataTables. You may consider server-side processing:
#> https://rstudio.github.io/DT/server.htmlwhere, for each gene and tumour_type,
N represents the total counts in the used dataset,
n is the count for the specific combination of
k and m.
1.2 User-defined priors
The user who may want to leverage priors obtained in a different way
(e.g. from other datasets or for a specific gene or tumour type not
included in priors_pcawg_hmf), can easily do that by
creating a similar data table.
For example:
my_priors = expand.grid(k=1:8, m = 1:8) %>%
dplyr::mutate(
gene = 'my_gene',
tumor_type = 'my_tumor_type') %>%
dplyr::filter(m<=k)
my_priors$n = rnorm(n = nrow(my_priors), mean = 50, sd = 10)
my_priors$N = sum(my_priors$n)The only requirement is that n is a positive number and
N is the sum of the values of n for a gene and
tumour type pair.
1.3 Visualising priors
The prior distribution used in a fit can be visualised a posteriori
using the internal plotting function plot_prior. We can
plot the prior distribution specific to a gene and tumour type used in
the example classified MSK-MET data.
For example:
plot_prior(x = priors_pcawg_hmf,
gene = 'KRAS',
tumor_type = 'PAAD')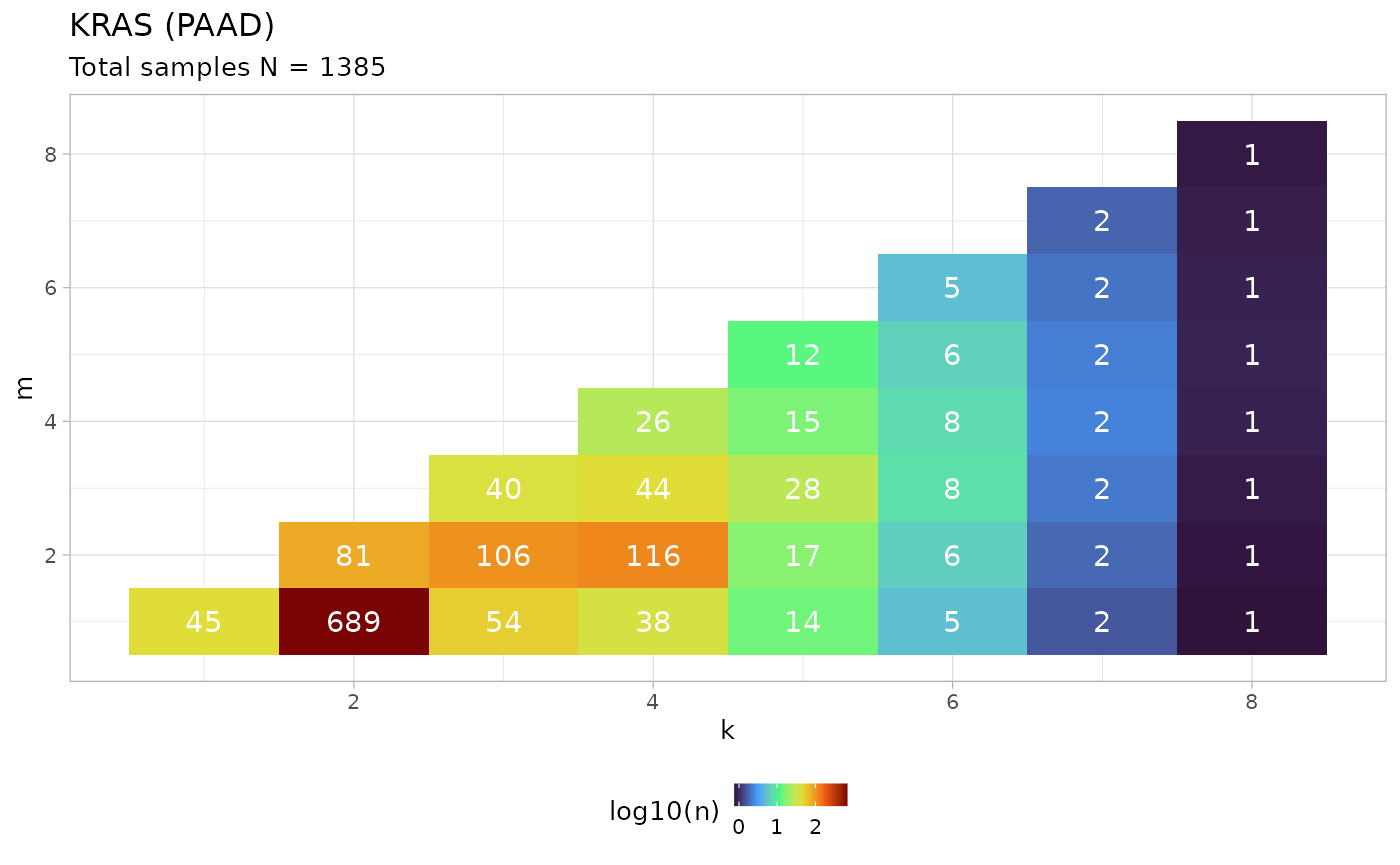
This example shows that, from the analysis of PCAWG and HMF datasets, KRAS mutations in pancreatic adenocarcinoma (PAAD) are most frequently found with configurations ( samples), i.e. in heterozygous diploid configurations. A relatively high frequency is also found for configurations with gain of the mutant copy: (CNLOH, samples), (mutant gain in trisomy, samples) and (mutant gain in tetrasomy, samples).