
Visualize classification results for individual observations.
Source:R/plot_classification.R
plot_classification.RdVisualize classification results for individual observations.
Examples
# First load example classified data
data(MSK_classified)
# Plot classification results for a specific sample
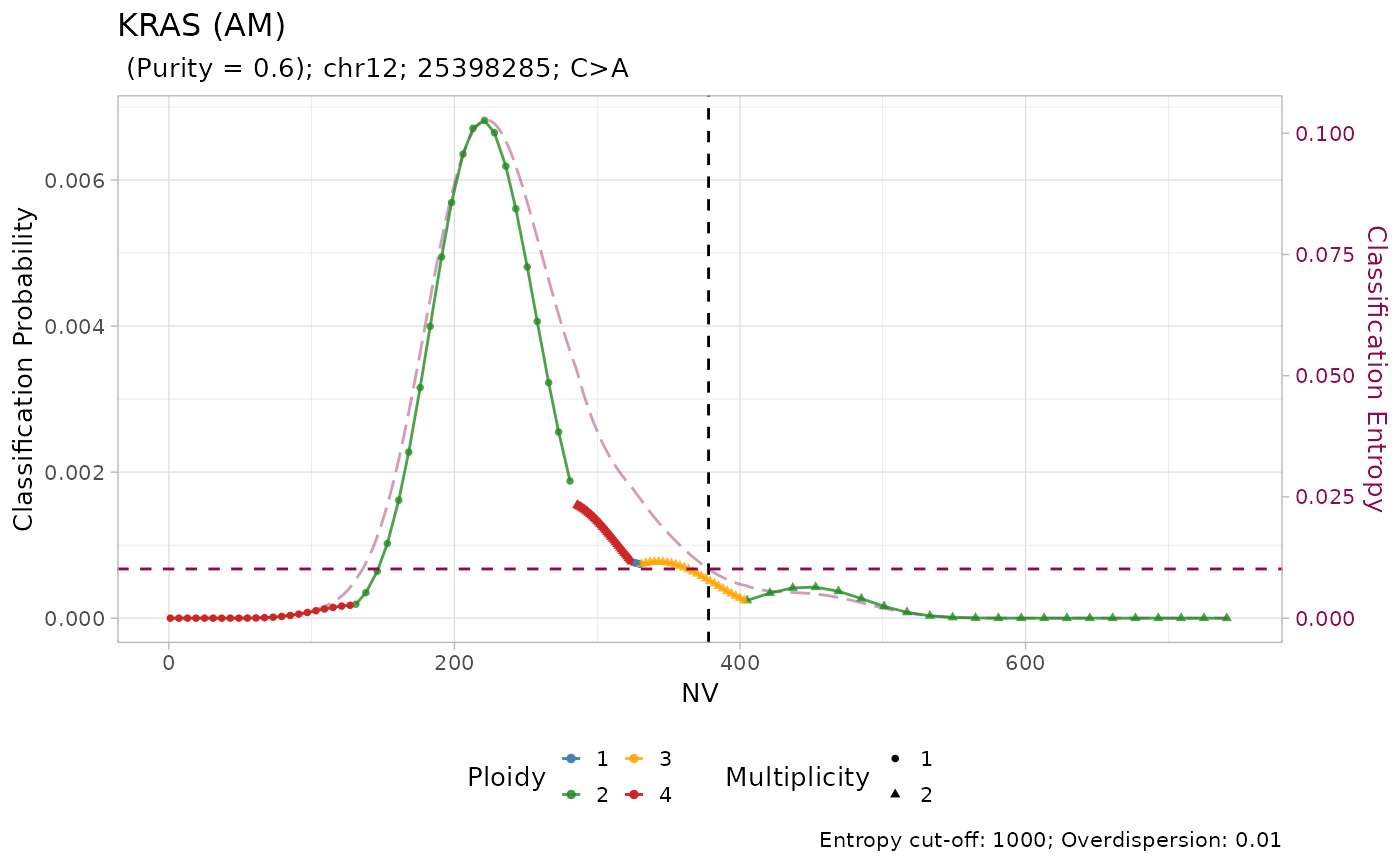
plot_classification(x = MSK_classified, sample = 'P-0002081-T01-IM3')
#> → No LUAD-specific prior probability specified for KRAS
#> → Using a pan-cancer prior
#> → No LUAD-specific prior probability specified for KRAS
#> → Using a pan-cancer prior
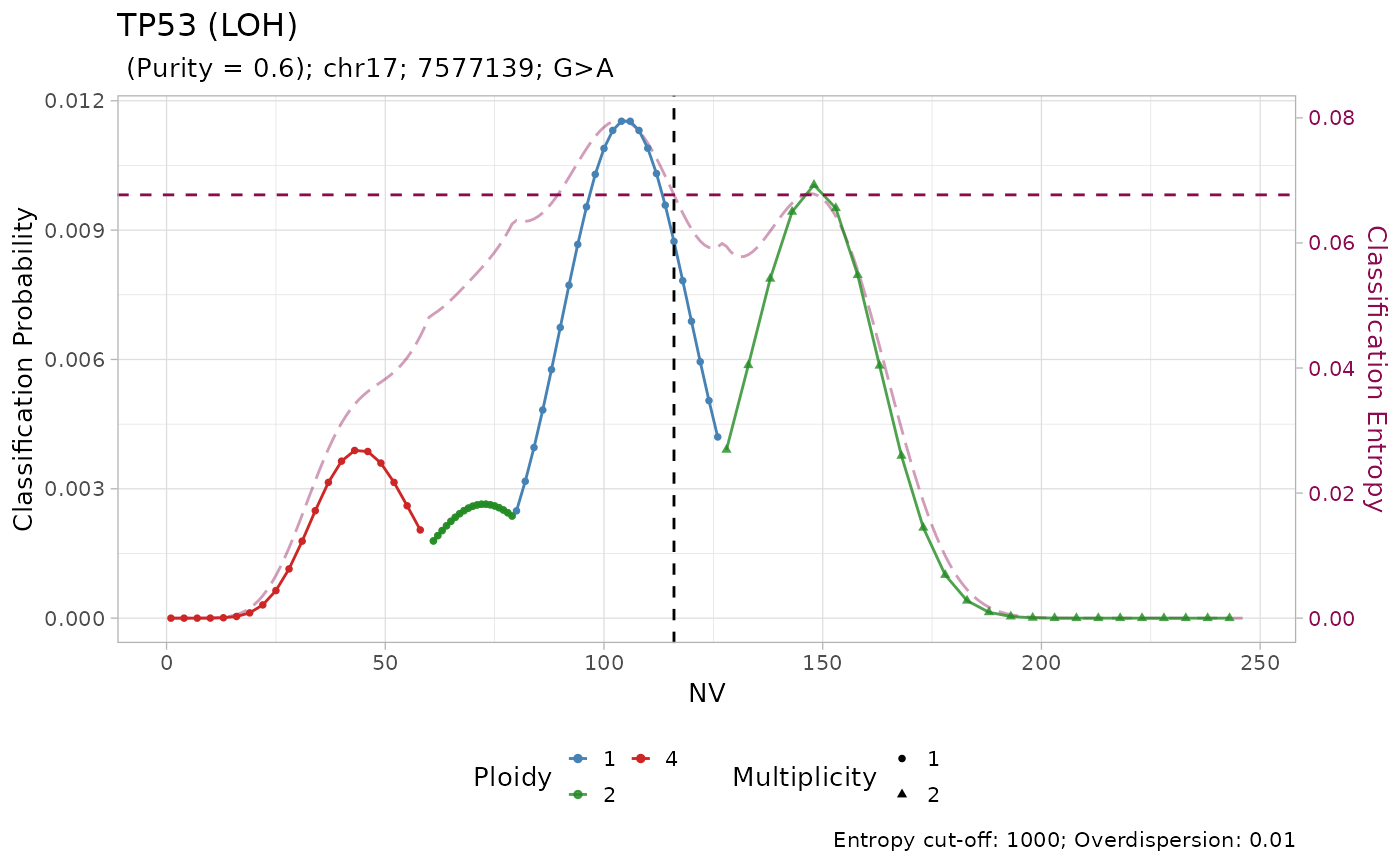
#> → No LUAD-specific prior probability specified for TP53
#> → Using a pan-cancer prior
#> → No LUAD-specific prior probability specified for TP53
#> → Using a pan-cancer prior
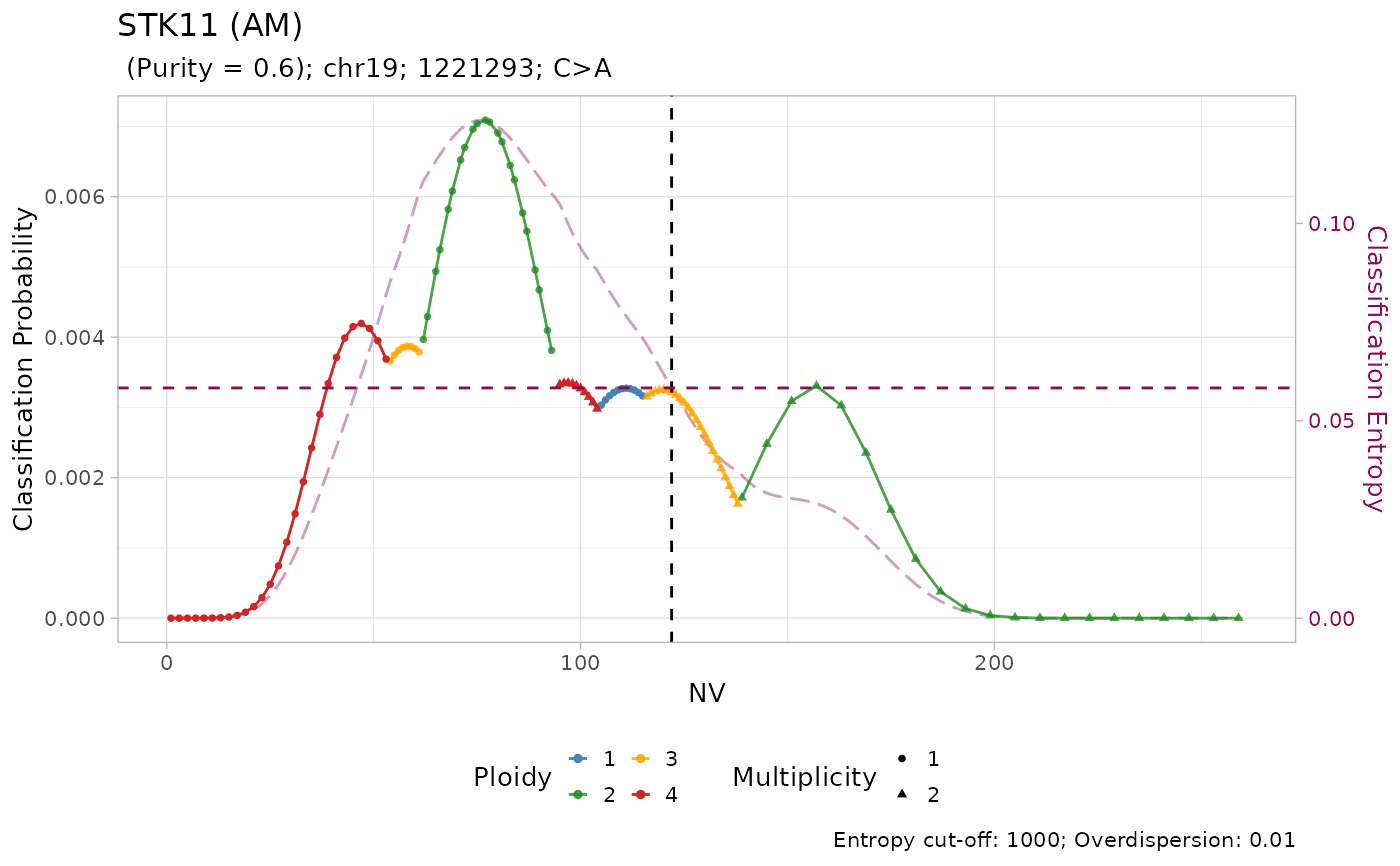
#> → No LUAD-specific prior probability specified for STK11
#> → Using a pan-cancer prior
#> → No LUAD-specific prior probability specified for STK11
#> → Using a pan-cancer prior
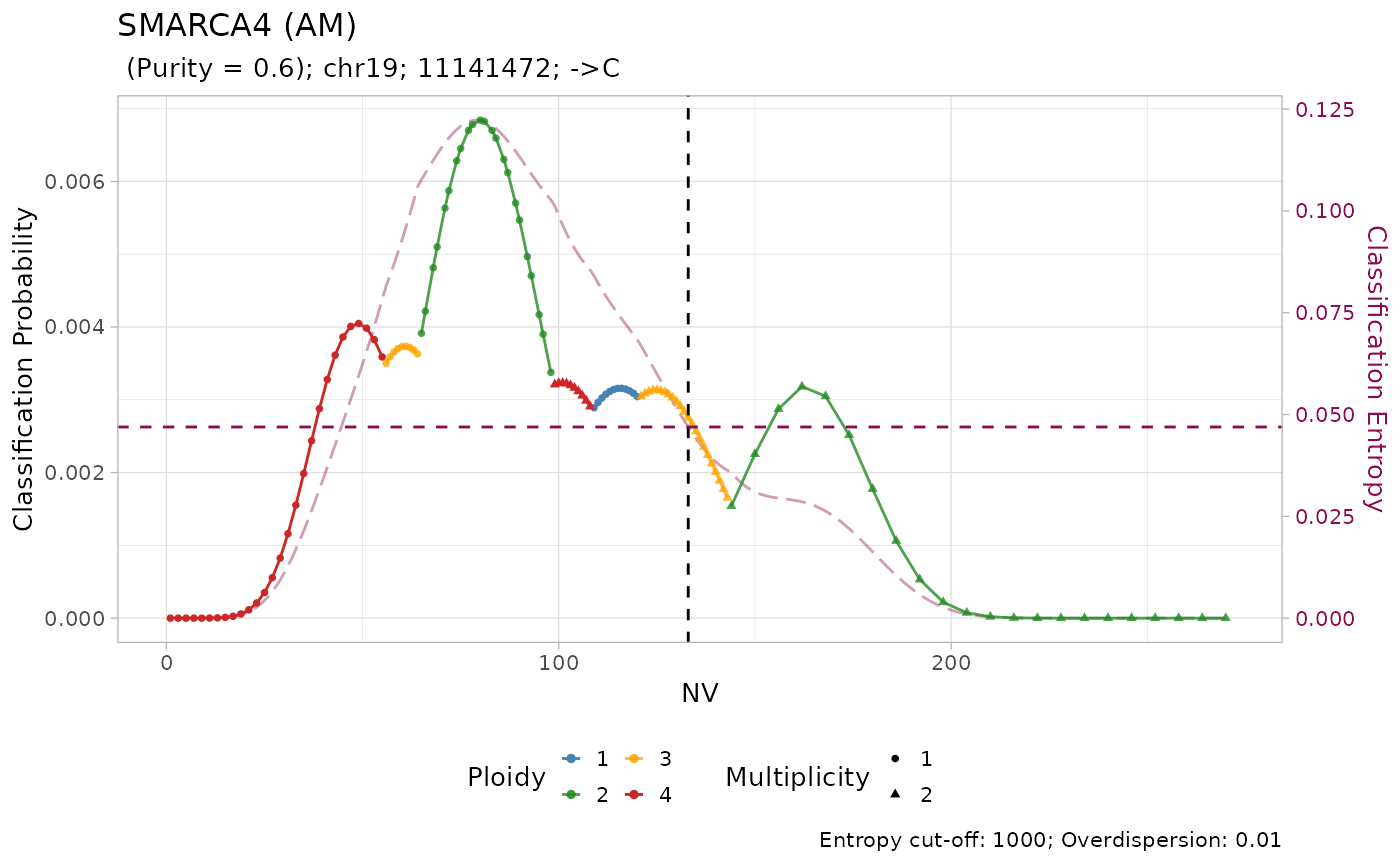
#> → No LUAD-specific prior probability specified for SMARCA4
#> → Using a pan-cancer prior
#> → No LUAD-specific prior probability specified for SMARCA4
#> → Using a pan-cancer prior
#> [[1]]
 #>
#> [[2]]
#>
#> [[2]]
 #>
#> [[3]]
#>
#> [[3]]
 #>
#> [[4]]
#>
#> [[4]]
 #>
#>