Title
Arguments
- cut_lfc
Value
Examples
x = Rcongas::congas_example
print(x)
#> ── [ Rcongas ] ────────────────────────────────────────────────────────────────
#>
#> → Data: 503 cells with 8564 genes, aggregated in 70 segments.
#>
#> ℹ Clusters: k = 2, model with AIC = 577427.35.
#>
#> ● Cluster c1, n = 379 [75.35% of total cells].
#> ● Cluster c2, n = 124 [24.65% of total cells].
#>
#> ── CNA highlights (alpha = 0.05)
#>
#> ✔ c1 CNA(s): [chr14:93300001:105450000] vs c2, [chr15:67050001:102600000] vs c2, [chr16:1:3750000] vs c2, and [chr18:32400001:55950000] vs c2
#>
#>
#> ── Differential Expression DESeq2 (1 vs 2)
#> ✔ 212 DE genes (alpha = 0.01, |lfc| > 0.25).
# Shows all data available
plot_DE_volcano(x)
 # More stringent parameters, annotate more genes
plot_DE_volcano(x, cut_pvalue = 1e-5, annotate_top = 10)
# More stringent parameters, annotate more genes
plot_DE_volcano(x, cut_pvalue = 1e-5, annotate_top = 10)
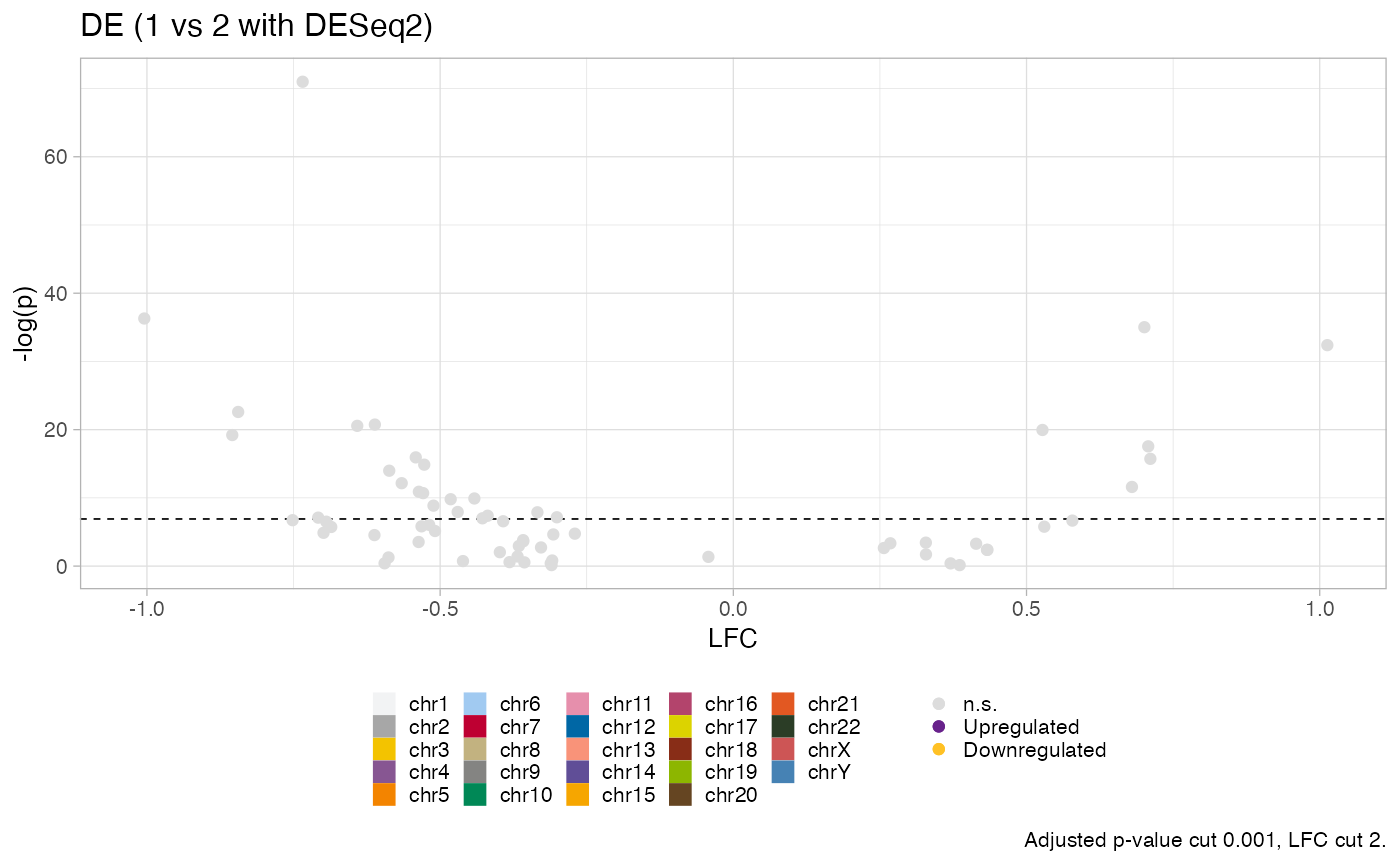
 # More stringent parameters and resitrict chromosome
plot_DE_volcano(x, cut_lfc = 2, chromosomes = c("chr1", "chr2"))
#> Warning: Removed 2 rows containing missing values (geom_vline).
# More stringent parameters and resitrict chromosome
plot_DE_volcano(x, cut_lfc = 2, chromosomes = c("chr1", "chr2"))
#> Warning: Removed 2 rows containing missing values (geom_vline).